













| General Information: |
|
| Name(s) found: |
SYFB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 598 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
phenylalanine-tRNA ligase complex [ISS] |
| Biological Process: |
phenylalanyl-tRNA aminoacylation
[IEA][ISS]
translation [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
magnesium ion binding [IEA] ATP binding [IEA] RNA binding [IEA] phenylalanine-tRNA ligase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTISVGRDR LFAALGESYT QEKFEELCFS FGIELDDVTT EKAIIRKEKH IDEEADDDEE 60
61 IIYKIEIPAN RPDLLCLEGL AQSLRVFIEK QEIPTYTLAD ISKDKILQMN VKPETSKIRP 120
121 FVVCAVLRGV TFDEARYNSF IDLQDKLHQN ICRRRSLVAI GTHDLDTLQG PFTYEALPPT 180
181 DINFVPLKQT KSFRADELIE FYKSDMKLKK FLHIIENSPV FPVLYDSKRT VLSLPPIING 240
241 AHSAITLQTK NVFIECTATD LTKAKIVLNT MVTTFSEFCA RKFEIEPVEV TYDDGKSYIY 300
301 PDLAVYDMEV PLSFITDSIG VSLKVEQVTS LLTRMQLQAE QAKSSDNQCA IKVHVPPSRS 360
361 DVLHPCDVME DVAIAYGFNN IPTRKPASIK PLTLNELTDL LRIEIAMCVY TEVVTWLLCS 420
421 HKENFAMLNR EDVNSAVIVG NPRSADFEAM RRALMPGLLK TVGHNNKYPK PIKIFEISDV 480
481 VMLDESKDVG ASNRRHLAAL YCGATSGFEL IHGLVDRIME VMAIPFLTIH ENNVPINEKD 540
541 GYYVKLSQEP EFLPGRQASI IVRGKHIGNF GIVHPEVLNN FDIPDPCSYL ELDIEAIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
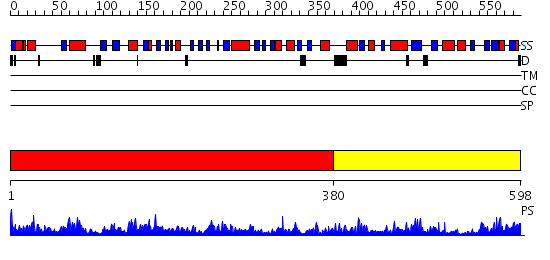
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..379] | 65.522879 | No description for 2cxiA was found. | |
| 2 | View Details | [380..598] | 45.30103 | Phenyl-tRNA synthetase (PheRS) alpha subunit, PheS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)