













| General Information: |
|
| Name(s) found: |
PHOT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 996 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
vacuole [IDA] internal side of plasma membrane [IDA] |
| Biological Process: |
chloroplast accumulation movement
[IMP]
chloroplast avoidance movement [IMP] negative regulation of anion channel activity by blue light [IMP] phototropism [IMP] regulation of stomatal movement [IMP] protein amino acid autophosphorylation [IDA][TAS] response to blue light [IGI] regulation of proton transport [IGI] |
| Molecular Function: |
protein binding
[IPI]
FMN binding [TAS] blue light photoreceptor activity [IDA] protein serine/threonine kinase activity [IDA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPTEKPSTK PSSRTLPRDT RGSLEVFNPS TQLTRPDNPV FRPEPPAWQN LSDPRGTSPQ 60
61 PRPQQEPAPS NPVRSDQEIA VTTSWMALKD PSPETISKKT ITAEKPQKSA VAAEQRAAEW 120
121 GLVLKTDTKT GKPQGVGVRN SGGTENDPNG KKTTSQRNSQ NSCRSSGEMS DGDVPGGRSG 180
181 IPRVSEDLKD ALSTFQQTFV VSDATKPDYP IMYASAGFFN MTGYTSKEVV GRNCRFLQGS 240
241 GTDADELAKI RETLAAGNNY CGRILNYKKD GTSFWNLLTI APIKDESGKV LKFIGMQVEV 300
301 SKHTEGAKEK ALRPNGLPES LIRYDARQKD MATNSVTELV EAVKRPRALS ESTNLHPFMT 360
361 KSESDELPKK PARRMSENVV PSGRRNSGGG RRNSMQRINE IPEKKSRKSS LSFMGIKKKS 420
421 ESLDESIDDG FIEYGEEDDE ISDRDERPES VDDKVRQKEM RKGIDLATTL ERIEKNFVIT 480
481 DPRLPDNPII FASDSFLELT EYSREEILGR NCRFLQGPET DLTTVKKIRN AIDNQTEVTV 540
541 QLINYTKSGK KFWNIFHLQP MRDQKGEVQY FIGVQLDGSK HVEPVRNVIE ETAVKEGEDL 600
601 VKKTAVNIDE AVRELPDANM TPEDLWANHS KVVHCKPHRK DSPPWIAIQK VLESGEPIGL 660
661 KHFKPVKPLG SGDTGSVHLV ELVGTDQLFA MKAMDKAVML NRNKVHRARA EREILDLLDH 720
721 PFLPALYASF QTKTHICLIT DYYPGGELFM LLDRQPRKVL KEDAVRFYAA QVVVALEYLH 780
781 CQGIIYRDLK PENVLIQGNG DISLSDFDLS CLTSCKPQLL IPSIDEKKKK KQQKSQQTPI 840
841 FMAEPMRASN SFVGTEEYIA PEIISGAGHT SAVDWWALGI LMYEMLYGYT PFRGKTRQKT 900
901 FTNVLQKDLK FPASIPASLQ VKQLIFRLLQ RDPKKRLGCF EGANEVKQHS FFKGINWALI 960
961 RCTNPPELET PIFSGEAENG EKVVDPELED LQTNVF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
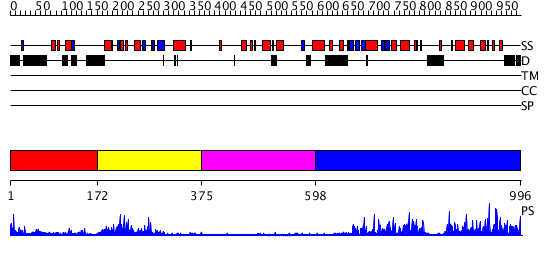
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [172..374] | 25.154902 | No description for 2z6cA was found. | |
| 3 | View Details | [375..597] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [598..996] | 80.39794 | No description for 2gnfA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)