













| General Information: |
|
| Name(s) found: |
NIA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 917 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
plasma membrane [IDA] |
| Biological Process: |
response to symbiotic fungus
[IEP]
nitric oxide biosynthetic process [IMP] response to light stimulus [IMP] nitrate assimilation [IMP] |
| Molecular Function: |
nitrate reductase activity
[IDA][ISS]
nitrate reductase (NADH) activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAASVDNRQY ARLEPGLNGV VRSYKPPVPG RSDSPKAHQN QTTNQTVFLK PAKVHDDDED 60
61 VSSEDENETH NSNAVYYKEM IRKSNAELEP SVLDPRDEYT ADSWIERNPS MVRLTGKHPF 120
121 NSEAPLNRLM HHGFITPVPL HYVRNHGHVP KAQWAEWTVE VTGFVKRPMK FTMDQLVSEF 180
181 AYREFAATLV CAGNRRKEQN MVKKSKGFNW GSAGVSTSVW RGVPLCDVLR RCGIFSRKGG 240
241 ALNVCFEGSE DLPGGAGTAG SKYGTSIKKE YAMDPSRDII LAYMQNGEYL TPDHGFPVRI 300
301 IIPGFIGGRM VKWLKRIIVT TKESDNFYHF KDNRVLPSLV DAELADEEGW WYKPEYIINE 360
361 LNINSVITTP CHEEILPINA FTTQRPYTLK GYAYSGGGKK VTRVEVTVDG GETWNVCALD 420
421 HQEKPNKYGK FWCWCFWSLE VEVLDLLSAK EIAVRAWDET LNTQPEKMIW NLMGMMNNCW 480
481 FRVKTNVCKP HKGEIGIVFE HPTLPGNESG GWMAKERHLE KSADAPPSLK KSVSTPFMNT 540
541 TAKMYSMSEV KKHNSADSCW IIVHGHIYDC TRFLMDHPGG SDSILINAGT DCTEEFEAIH 600
601 SDKAKKMLED YRIGELITTG YSSDSSSPNN SVHGSSAVFS LLAPIGEATP VRNLALVNPR 660
661 AKVPVQLVEK TSISHDVRKF RFALPVEDMV LGLPVGKHIF LCATINDKLC LRAYTPSSTV 720
721 DVVGYFELVV KIYFGGVHPR FPNGGLMSQY LDSLPIGSTL EIKGPLGHVE YLGKGSFTVH 780
781 GKPKFADKLA MLAGGTGITP VYQIIQAILK DPEDETEMYV IYANRTEEDI LLREELDGWA 840
841 EQYPDRLKVW YVVESAKEGW AYSTGFISEA IMREHIPDGL DGSALAMACG PPPMIQFAVQ 900
901 PNLEKMQYNI KEDFLIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
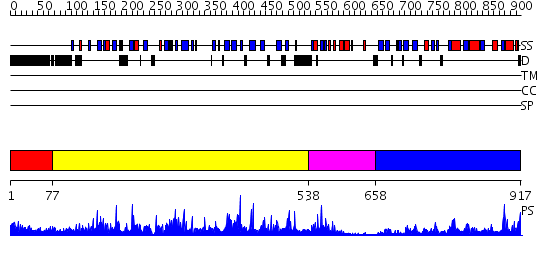
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..537] | 136.0 | crystal structure of the Molybdenum-containing nitrate reducing fragment of Pichia angusta assimilatory nitrate reductase | |
| 3 | View Details | [538..657] | 14.522879 | Cytochrome b5 | |
| 4 | View Details | [658..917] | 62.39794 | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)