













| General Information: |
|
| Name(s) found: |
DEGP8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 448 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
thylakoid lumen [IDA] chloroplast thylakoid lumen [IDA] thylakoid [IDA] |
| Biological Process: |
proteolysis
[IDA][ISS]
photosystem II repair [IMP] |
| Molecular Function: |
serine-type peptidase activity
[ISS]
peptidase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQVIASFCSK PNENEFVGRR QLLSSVCSKI SQGDVVSHPP VSSVKVTQDW KSNLHELAVK 60
61 SVPSTTRRIL LTSLFMNLCF NPSRYLSALA LGDPSVATVE DVSPTVFPAG PLFPTEGRIV 120
121 QLFEKNTYSV VNIFDVTLRP QLKMTGVVEI PEGNGSGVVW DGQGYIVTNY HVIGNALSRN 180
181 PSPGDVVGRV NILASDGVQK NFEGKLVGAD RAKDLAVLKV DAPETLLKPI KVGQSNSLKV 240
241 GQQCLAIGNP FGFDHTLTVG VISGLNRDIF SQTGVTIGGG IQTDAAINPG NSGGPLLDSK 300
301 GNLIGINTAI FTQTGTSAGV GFAIPSSTVL KIVPQLIQFS KVLRAGINIE LAPDPVANQL 360
361 NVRNGALVLQ VPGKSLAEKA GLHPTSRGFA GNIVLGDIIV AVDDKPVKNK AELMKILDEY 420
421 SVGDKVTLKI KRGNEDLELK ISLEEKSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
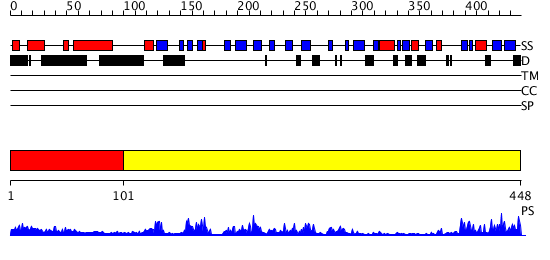
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [101..448] | 65.522879 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)