













| General Information: |
|
| Name(s) found: |
PRP43_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 729 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
chloroplast envelope [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] ATP binding [IEA] RNA helicase activity [ISS] nucleoside-triphosphatase activity [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTERKRKVS LFDVMEDPSL SSKNTKSNGL GLAAAAGGGS NLINKWNGKA YSQRYFEILE 60
61 KRRDLPVWLQ KDDFLNTLNS NQTLILVGET GSGKTTQIPQ FVLDAVVADN SDKGRKWLVG 120
121 CTQPRRVAAM SVSRRVADEM DVSIGEEVGY SIRFEDCTSS RTMLKYLTDG MLLREAMADP 180
181 LLERYKVIIL DEAHERTLAT DVLFGLLKEV LRNRPDLKLV VMSATLEAEK FQEYFSGAPL 240
241 MKVPGRLHPV EIFYTQEPER DYLEAAIRTV VQIHMCEPPG DILVFLTGEE EIEDACRKIN 300
301 KEVSNLGDQV GPVKVVPLYS TLPPAMQQKI FDPAPVPLTE GGPAGRKIVV STNIAETSLT 360
361 IDGIVYVIDP GFAKQKVYNP RIRVESLLVS PISKASAHQR SGRAGRTRPG KCFRLYTEKS 420
421 FNNDLQPQTY PEILRSNLAN TVLTLKKLGI DDLVHFDFMD PPAPETLMRA LEVLNYLGAL 480
481 DDEGNLTKTG EIMSEFPLDP QMSKMLIVSP EFNCSNEILS VSAMLSVPNC FVRPREAQKA 540
541 ADEAKARFGH IDGDHLTLLN VYHAYKQNNE DPNWCFENFV NNRAMKSADN VRQQLVRIMS 600
601 RFNLKMCSTD FNSRDYYVNI RKAMLAGYFM QVAHLERTGH YLTVKDNQVV HLHPSNCLDH 660
661 KPEWVIYNEY VLTTRNFIRT VTDIRGEWLV DVAQHYYDLS NFPNCEAKRA LEKLYKKRER 720
721 EKNESKNRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
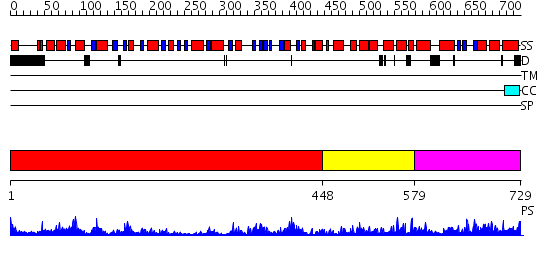
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..447] | 66.30103 | No description for 2hyiC was found. | |
| 2 | View Details | [448..578] | 1.49 | No description for 2v8oA was found. | |
| 3 | View Details | [579..729] | 26.920819 | No description for PF07717.7 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)