













| General Information: |
|
| Name(s) found: |
DING_BACSU
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Bacillus subtilis |
| Length: | 931 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKQRFVVID VETTGNSPKK GDKIIQIAAV VIENGQITER FSKYINPNKS IPAFIEQLTG 60
61 ISNQMVENEQ PFEAVAEEVF QLLDGAYFVA HNIHFDLGFV KYELHKAGFQ LPDCEVLDTV 120
121 ELSRIVFPGF EGYKLTELSE ELQLRHDQPH RADSDAEVTG LIFLEILEKL RQLPYPTLKQ 180
181 LRRLSQHFIS DLTHLLDMFI NENRHTEIPG YTRFSSFSVR EPEAIDVRIN EDENFSFEIE 240
241 SWEAGNEKAL SELMPGYEKR DGQMMMMREV ADAFANREHA LIEAPPGIGK TIGYLIPAAL 300
301 FAKKSKKPVI ISTYSTLLQQ QILTKDLPIV QDLFPFPVTA AILKGQSHYL CLYKFEQVLH 360
361 EEDDNYDAVL TKAQLLVWLT ETNTGDVAEL NLPSGGKLLW DRLAYDDDSY KRSRSEHVIG 420
421 FYERAKQIAM RSDLVITNHS LLLTDEGSHK KRLPESGTFI IDEAHHFERA ASEHLGKRAT 480
481 YIELHTKLSR IGTLKEQGLL KKMRQLFQRN SLPVDSFFEL EEWLQHVQAE SDAFFSSVHS 540
541 FVKRRKPKED LNRLVFKVNK ESQDKSWSIL TDGAERLCSM LTHLQQLFEA QSSLMEKHLK 600
601 GMKSKTVFLA DEYQRSMKGL QHYCQTLQKL FFGSDDDEAV WIEIDAKGAK NAVAIYAQPL 660
661 EPGELLADQF FARKNSVVLT SATLTVEGSF QFMIERLGLS DFFPRTMRIE SPFSYDERMQ 720
721 VMIPKEMKSI QDTGQPEFIQ DTARYIELMA KEKQPKILVL FTSHDMLKKV HQELKHNMSA 780
781 SGIQLLAQGI TGGSPGKLMK TFKTSNQAIL LGTNHFWEGV DFPGDELTTV MIVRLPFRSP 840
841 DHPLHAAKCE LARKKGKNPF QTVSLPEAVL TFRQGIGRLL RSAGDKGTII ILDRRIKTAG 900
901 YGRLFLDALP TTSVSEMTDS ELEAYVAGEK E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
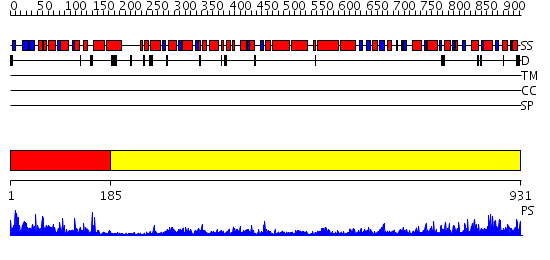
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 42.09691 | No description for 2guiA was found. | |
| 2 | View Details | [185..931] | 98.39794 | No description for 2fdcA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)