













| General Information: |
|
| Name(s) found: |
CE36100
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
cellular amino acid metabolic process
[IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA cystathionine beta-lyase activity [IEA transferase activity, transferring alkyl or aryl (other than methyl) groups [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRTDNLSDL ASARDYWRTD KYISFVITFQ FYLLLDTMSD SFNWPAGNNE ARERLGEKFE 60
61 SLHLDSRIST SHAKPLSNAD PVVVPIYHSS TYRFKTVDQF NEDNHGANFV YRRCGNPTTE 120
121 NVEVVINEIE GGAGSLLYNS GLAAISAVFL EFLSSGAHMI VMNPIYSGTS SFINETLARF 180
181 GVEITSVDVE KEKDFAGAVE KAIRPNTKMI YFESIANPTM AVPDILGTIE VAKKYKILTC 240
241 LDATFSSPYN IQPLKLGADI SLHSCSKYIG GHTDVIAGVV TVSSYDNWKK LKLQQLTTGS 300
301 SLSPYDAALL TRGLKTLGLR VDRISENAQK TAEFLESHPK VERVFYPGLP SHPQHQYAKQ 360
361 VMKQFAGMIA FDVGTAENAI KLVESLKLII HAVSLGGTES LIEHPLSMSH GKHLLRYLDG 420
421 PTVAPGLLRF SVGIENVEDI IGDLNDALEK L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
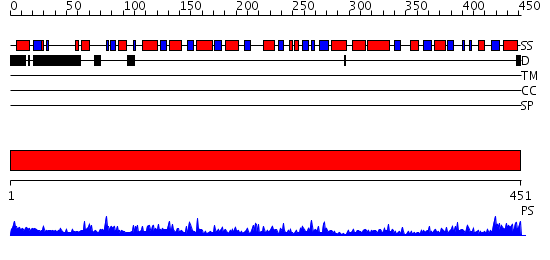
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 109.0 | Crystal structure of Citrobacter Freundii L-methionine-lyase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)