













| General Information: |
|
| Name(s) found: |
nhr-34 /
CE35565
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 631 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDYCIFNIS WMSDRFGCET GNEDVQECID KSPNGSPTTS TTHRRQNSSS GLEPARKRPK 60
61 LSPPVLTAMP LDLTDAVSPN ESQACTSSLK GVVTAAGAAQ MSVADRMTSS SVAPPLCIVP 120
121 RSVEFINPKT EDMSMDIPMG NVQSEFTGLK CRVCGDSRAG RHYGTIACNG CKGFFRRSIW 180
181 EQRDYVCRFG GKCLVVQEYR NRCRACRLRK CFTVGMDARA VQSERDKHKK NPKDSNNEGS 240
241 TSPQYPTAST PISIPSTSTS QTPTSSVNSY NFQNIPGIVS RSFSENLIMR DNSVPVMETS 300
301 QSAALSHVPL VRYLIDLEKA TDNLIDENCD FMSMEFDQLC RVDVTIEAAF RQPGVVAKRT 360
361 PPRWLALERL TTLEDVHIAW CRSFVLCIDY AKIMKDYQEL SPTDQFTLLR NRVISVNWLC 420
421 HTYKTFKAGC DGVALVNGSW YPRDKELQKQ LDPGCNHYFR ILSEHLMEDL VIPMREMDMD 480
481 EGEFVILKAL ILFRAHRRLS EEGRAHIKRV RDKYIEALYQ HVQHQHRHFS SVQTSMRISK 540
541 ILLLLPSIEH LSQQEDDNVQ FLALFNLANL NGLPYELHSS IKQHIPNGDD SDDTQVNEVT 600
601 SNNDGPRSSE SSHTPQSVST SQFLEFKPSL H |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
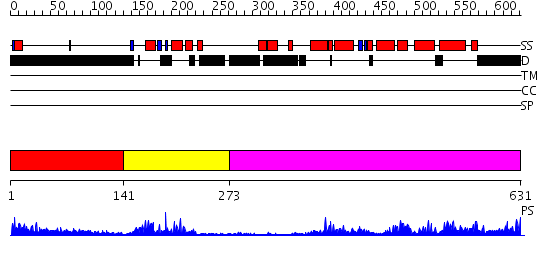
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 8.361996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [141..272] | 26.39794 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 3 | View Details | [273..631] | 56.154902 | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)