













| General Information: |
|
| Name(s) found: |
CE33003
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
protein modification process
[IEA ubiquitin cycle [IEA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVQTVVHGY PPNTDNETEL LRVRLLRGTK LGKRDLLGVC SPYCMIRLEN SNGDTVDEVQ 60
61 VETKKKTRNP TWNSVFTFRV TRACQMKILV YDENRLRKDV LMGFVSRTLN DGLITTQAPN 120
121 PEEYALQSGT ASKGKSIGSL FLSFHFMPSS SDNSPSDSTA DLQQTSSGMP QGWEEREDGN 180
181 GRTVYVNHAL RTTQFTPPES VSNGNVDAIT EQSVIEETRR RRDNYEHRSQ VTDEPSDTTI 240
241 VSNELTAIDD AMRANFERGG GHVEEEEDEL RLPDGWDMQV APNGRTFFID HRTKTTTWTD 300
301 PRPGAATRVP LLRGKTDDEI GALPAGWEQR VHADGRVFFI DHNRRRTQWE DPRFENENIA 360
361 GPAVPYSRDY KRKVEYLRSR LPKPNSNSGK CDMVVHRDTL FEDSYRHIMD KKDYDLRNKL 420
421 WIEFFGETGL DYGGVTREWF FLLSHQIFNP YYGLFEYSAT DNYTLQINPH SEACNPEHLS 480
481 YFHFIGRIIG MAIYHGKLLD AFFIRPFYKM MLGKKITLFD MESVDNEYYN SLIYVKDNDP 540
541 ADLELTFSLD DSIFGETQNI ELIPNGANVP VTEDNKEEYI EAVVPSNLLR LFDANELELL 600
601 MCGLQKIDVK DWKANTIYKG GYGPSSQVVH NFWKCILSFD NEMRARVLQF VSGTSRVPMN 660
661 GFRELYGSNG LQKFTIERWG SADMLPRAHT CFNRLDLPPY TTFKELKSKL LTAIENSEIF 720
721 SGVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
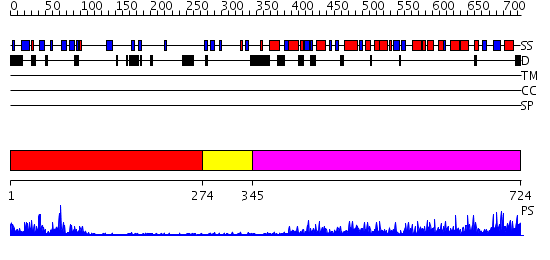
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..273] | 33.30103 | Synaptogamin I | |
| 2 | View Details | [274..344] | 4.01 | Splicing factor prp40 | |
| 3 | View Details | [345..724] | 106.0 | No description for 2oniA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)