













| General Information: |
|
| Name(s) found: |
coq-6 /
CE30815
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular aromatic compound metabolic process
[IEA electron transport [IEA determination of adult lifespan [IMP ubiquinone biosynthetic process [IEA |
| Molecular Function: |
FAD binding
[IEA monooxygenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLPGGTIIC ARNASSYYDT VIVGGGMVGN AMACSLGANK SFQSKSVLLL DAGRSPSLAS 60
61 FKPGAPFNNR VVATSPTSID TFKKLGVWDQ INSHRTKKVN RLFVFDSCST SEIEFERGQQ 120
121 EEVAFIIEND LIVGSLYEKL AEYKNVDVKT GAKVEDCSIP NALENMATIK LENGDVIETS 180
181 LLIGADGVNS KVRHASNLDY TTFNYNQHGL VAIVNIETAN GKNETAWQRF TTLGPVALLP 240
241 LSDTVSGLTW STSPEEAQRL KQLPSDQFVD ELNSALFSQN NQIPLVNQTI FALNRMNPFR 300
301 TETFGRKAEG TTPPHVITVQ DKSRASFPLG FGNAHSYITT RCALIGDAAH RMHPLAGQGV 360
361 NLGWSDVQIL DKVLGDAVRE GADIGSITYL REYDSAAQKH NLPVMVSVDL LNRLYRTDAP 420
421 AIVAARAFGL NAFNSLGPVK NFLMNYLSAH R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
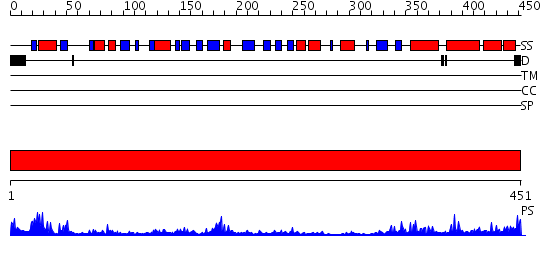
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 46.09691 | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 42 REPLACED BY LYS (R42K), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)