













| General Information: |
|
| Name(s) found: |
eri-1 /
CE30163
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA intracellular [IEA |
| Biological Process: |
reproductive process in a multicellular organism
[IMP meiotic chromosome segregation [IMP RNA interference [IMP RNA catabolic process [IDA |
| Molecular Function: |
nucleic acid binding
[IEA 3'-5'-exoribonuclease activity [IDA exonuclease activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSADEPSPED EKYLESLRDL LKISQEFDAS NAKQNDEPEK TAVEVESAET RTDESEKSID 60
61 IPREQQLLPS ERVEPLKSMV EPEYVKKVIR QMDTMTAEQL KQALMKIKVS TGGNKKTLRK 120
121 RVAQYYRKEN ALLNRKMEPN ADKTARFFDY LIAIDFECTC VEIIYDYPHE IIELPAVLID 180
181 VREMKIISEF RTYVRPVRNP KLSEFCMQFT KIAQETVDAA PYFREALQRL YTWMRKFNLG 240
241 QKNSRFAFVT DGPHDMWKFM QFQCLLSNIR MPHMFRSFIN IKKTFKEKFN GLIKGNGKSG 300
301 IENMLERLDL SFVGNKHSGL DDATNIAAIA IQMMKLKIEL RINQKCSYKE NQRSAARKDE 360
361 ERELEDAANV DLTSVDISRR DFQLWMRRLP LKLSSVTRRE FINEEYLDCD SCDDLTDDKN 420
421 DEAAFQEKMA IREYLENKQT EDFAKIAAER GIFKIGEIKS YQTARPIIED DDVDVESEEE 480
481 DYGTEFEMLE VVERMPPVSS TLHTEVDLDA VWERDGGSDS ERENLSNAPS LHEFPSSSTS 540
541 SPHATSEHVT SSSPLHIDDD VDRVLNAPPK NSLASSSNRS SF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
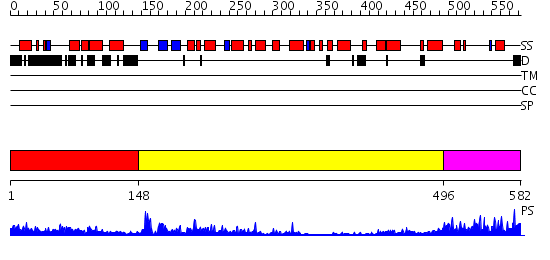
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 43.221849 | No description for 1zbuA was found. | |
| 2 | View Details | [148..495] | 23.0 | Exonuclease I | |
| 3 | View Details | [496..582] | 34.09691 | No description for 2hnhA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)