













| General Information: |
|
| Name(s) found: |
cep-1 /
CE29805
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 644 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA nucleoplasm [IDA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP meiotic chromosome segregation [IMP DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis [IGI chromosome segregation [IMP regulation of transcription [IDA response to stress [IMP |
| Molecular Function: |
DNA binding
[IDA transcription factor activity [IDA zinc ion binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPDDSQLSD ILKDARIPDS QDIGVNLTQN LSFDTVQKMI DGVFTPIFSQ GTEDSLEKDI 60
61 LKTPGISTIY NGILGNGEET KKRTPKISDA FEPDLNTSGD VFDSDKSEDG LMNDESYLSN 120
121 TTLSQVVLDS QKYEYLRVRT EEEQQLVIEK RARERFIRKS MKIAEETALS YENDGSRELS 180
181 ETMTQKVTQM DFTETNVPFD GNDESSNLAV RVQSDMNLNE DCEKWMEIDV LKQKVAKSSD 240
241 MAFAISSEHE KYLWTKMGCL VPIQVKWKLD KRHFNSNLSL RIRFVKYDKK ENVEYAIRNP 300
301 RSDVMKCRSH TEREQHFPFD SFFYIRNSEH EFSYSAEKGS TFTLIMYPGA VQANFDIIFM 360
361 CQEKCLDLDD RRKTMCLAVF LDDENGNEIL HAYIKQVRIV AYPRRDWKNF CEREDAKQKD 420
421 FRFPELPAYK KASLESINIK QEVNLENMFN VTNTTAQMEP STSYSSPSNS NNRKRFLNEC 480
481 DSPNNDYTMM HRTPPVTGYA SRLHGCVPPI ETEHENCQSP SMKRSRCTNY SFRTLTLSTA 540
541 EYTKVVEFLA REAKVPRYTW VPTQVVSHIL PTEGLERFLT AIKAGHDSVL FNANGIYTMG 600
601 DMIREFEKHN DIFERIGIDS SKLSKYYEAF LSFYRIQEAM KLPK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
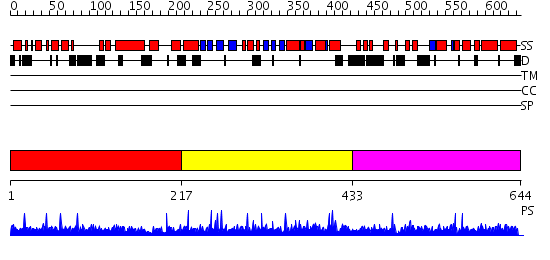
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [217..432] | 111.0 | Structural Differences in the DNA Binding Domains of Human p53 and its C. elegans Ortholog Cep-1: Structure of C. elegans Cep-1 | |
| 3 | View Details | [433..644] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)