













| General Information: |
|
| Name(s) found: |
pef-1 /
CE30662
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA cytoplasm [IEA neuronal cell body [IDA |
| Biological Process: |
detection of stimulus involved in sensory perception
[IEA protein amino acid dephosphorylation [IEA |
| Molecular Function: |
phosphoprotein phosphatase activity
[IEA bis(5'-nucleosyl)-tetraphosphatase (symmetrical) activity [IEA iron ion binding [IEA calcium ion binding [IEA manganese ion binding [IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEENGKGGV ENGRNSPLMS ALSHYAKPSL MDSEGETVKK MLEDTSPTNV DIDRNYKGPT 60
61 LSLPLDKPQV AKMIEAFKVN KVLHPKYVLM ILHEARKIFK AMPSVSRIST SISNQVTICG 120
121 DLHGKFDDLC IILYKNGYPS VDNPYIFNGD FVDRGGQSIE VLCVLFALVI VDPMSIYLNR 180
181 GNHEDHIMNL RYGFIKELST KYKDLSTPIT RLLEDVFSWL PIATIIDRDI FVVHGGISDQ 240
241 TEVSKLDKIP RHRFQSVLRP PVNKGMESEK ENSAVNVDEW KQMLDIMWSD PKQNKGCWPN 300
301 VFRGGGSYFG ADITASFLEK HGFRLLVRSH ECKFEGYEFS HNNTCLTVFS ASNYYETGSN 360
361 RGAYVKFIGK SKQPHFVQYM ASKTHRKSTL RERLGVVEES AVKELKEKLS SFHTDLQKEF 420
421 EIMDIEKSGK LPILKWSDCV ERITGLNLPW IALAPKVATL SEDGKYVMYK EDRRIAQVGG 480
481 THAQEKDIVE SLYRHKSTLE TLFRFMDKDN SGQVSMKEFI DACEVLGKYT KRPLQTDYIS 540
541 QIAESIDFNK DGFIDLNELL EAFRLVDRPL LR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
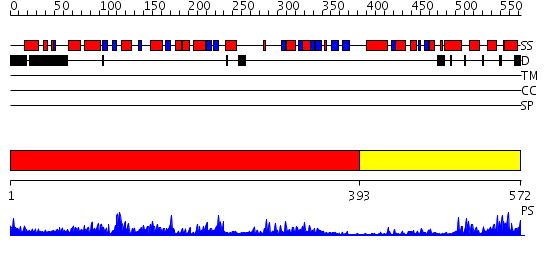
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..392] | 92.69897 | PP5 structure | |
| 2 | View Details | [393..572] | 29.69897 | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)