













| General Information: |
|
| Name(s) found: |
daf-16 /
CE28771
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 530 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to heat
[IEP dauer larval development [IGI insulin receptor signaling pathway [IDA determination of adult lifespan [IGI regulation of transcription, DNA-dependent [IEA positive regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
transcription factor activity
[IDA protein binding [IPI sequence-specific DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDSIDDDFP PEPRGRCYTW PMQQYIYQES SATIPHHHLN QHNNPYHPMH PHHQLPHMQQ 60
61 LPQPLLNLNM TTLTSSGSSV ASSIGGGAQC SPCASGSSTA ATNSSQQQQT VGQMLAASVP 120
121 CSSSGMTLGM SLNLSQGGGP MPAKKKRCRK KPTDQLAQKK PNPWGEESYS DIIAKALESA 180
181 PDGRLKLNEI YQWFSDNIPY FGERSSPEEA AGWKNSIRHN LSLHSRFMRI QNEGAGKSSW 240
241 WVINPDAKPG RNPRRTRERS NTIETTTKAQ LEKSRRGAKK RIKERALMGS LHSTLNGNSI 300
301 AGSIQTISHD LYDDDSMQGA FDNVPSSFRP RTQSNLSIPG SSSRVSPAIG SDIYDDLEFP 360
361 SWVGESVPAI PSDIVDRTDQ MRIDATTHIG GVQIKQESKP IKTEPIAPPP SYHELNSVRG 420
421 SCAQNPLLRN PIVPSTNFKP MPLPGAYGNY QNGGITPINW LSTSNSSPLP GIQSCGIVAA 480
481 QHTVASSSAL PIDLENLTLP DQPLMDTMDV DALIRHELSQ AGGQHIHFDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
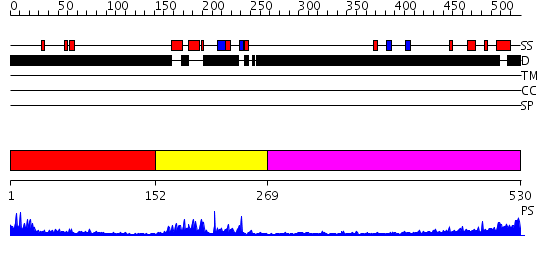
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 1.443994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [152..268] | 24.0 | Afx (Foxo4) | |
| 3 | View Details | [269..530] | 5.461996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)