













| General Information: |
|
| Name(s) found: |
fax-1 /
CE28464
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 419 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
locomotory behavior
[IMP neuron fate specification [IMP regulation of transcription, DNA-dependent [IEA regulation of forward locomotion [IMP regulation of backward locomotion [IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDEDEPLNF STSKATEESK EGILGVRSIF NTPLLFPPPM FNAGVISPHI AAALAMSFNQ 60
61 QRMNASVSPP LDHTTISVNS FPSMGSVKTD SPPTASSPTL CCAVCGDVSS GKHYGILACN 120
121 GCSGFFKRSV RRRLIYRCQA GTGNCVVDKA HRNQCQACRL KKCLNKGMNK DAVQNERQPR 180
181 NTATIRPALD MDPQNFFREY AGAVSAIMGH SNMMKREDSP SSASDGKTED EKKDSLQETT 240
241 MSQLESVLQW AQQFRLFTVL TNSEKRQIIL TQWPRLLCIS LCEQAEDVSF DDHLTSLMLK 300
301 FRRLDVSPAE FNCLKAITIF MKRELSLWRA GWDNRASIIT VYPAGERGAR LVAAALLLAE 360
361 HSVMGFGNCV IPLALVFSTK SRYVIQRHAI NSLPACVPGG TSAHPVLRCS MGSRREKVI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
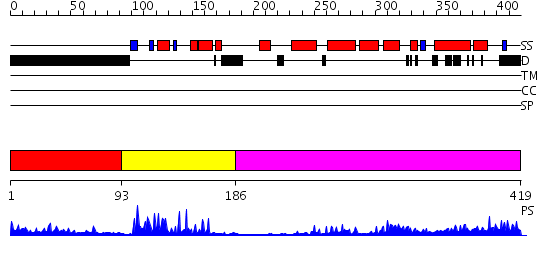
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 5.486997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [93..185] | 25.0 | Orphan nuclear receptor reverb | |
| 3 | View Details | [186..419] | 43.69897 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.40 |
Source: Reynolds et al. (2008)