













| General Information: |
|
| Name(s) found: |
sir-2.4 /
CE27656
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 299 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin silencing complex
[IEA |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA chromatin silencing [IEA |
| Molecular Function: |
DNA binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSAKYKTVS SKKRFFKHNS IIFLIQNFQI RDTETEIIEK LRTLYNHFVQ AKQTGKPIFV 60
61 LIGAGVSTGS KLPDFRGKQG VWTLQAEGKH AEGVDFQVAR PGVSHKSILA LHKAGYIKTI 120
121 ITQNVDGLDR KVGIPVEDLI EVHGNLFLEV CQSCFSEYVR EEIVMSVGLC PTGRNCEGNK 180
181 RTGRSCRGKL RDATLDWDTE ISLNHLDRIR KAWKQTSHLL CIGTSLEIIP MGSLPLDAKS 240
241 KGIKTTTINY QETAHEKIVE TAIHADVKLI LYSLCNALGV NVDLGDDLPD EVPIPLKIS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
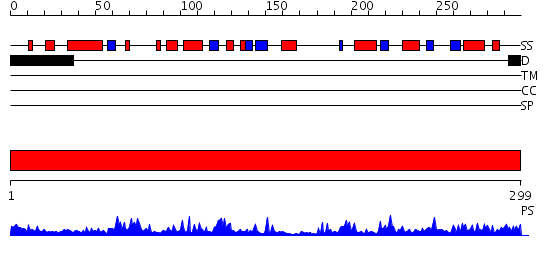
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 64.69897 | AF1676, Sir2 homolog (Sir2-AF1?) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)