













| General Information: |
|
| Name(s) found: |
par-4 /
CE27410
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 617 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cell cortex [IDA |
| Biological Process: |
asymmetric protein localization involved in cell fate determination
[IMP cell differentiation [IMP establishment of cell polarity [IMP asymmetric cell division [IMP protein amino acid phosphorylation [IEA |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAPSTSSGA QSKLLMPGDD EADEDHQNRG DPNLQQKQKI QLNVDPDYDD DEDDDCFIDG 60
61 CEASAPITRE LVDGAIERRS KDRNVKMSIG VYDEYDDDDD DEEETEEDQR RRFVEGIRNI 120
121 RHKQQESFDL EEHPIPVESE AMRQFINQQV NNAMMFNQDN SEFQHIEFEP IVKQKGPKII 180
181 EGYMWGGQIG TGSYGKVKEC IDMYTLTRRA VKIMKYDKLR KITNGWENIR SEMSILRRMN 240
241 HRNVIKLIEI FNIPAKGKVY MVFEYCIGSV QQLLDMEPAR RLTIGESHAI FIELCQGLNY 300
301 LHSKRVSHKD IKPGNLLVSI DFTVKICDFG VAEQINLFQR DGRCTKVNGT PKFQPPECIY 360
361 GNHDFFDGYK ADMWSAGVTL YNLVSGKYPF EKPVLLKLYE CIGTEPLQMP TNVQLTKDLQ 420
421 DLLTKLLEKD FNERPTCLET MIHPWFLSTF PEDQGLGRIM ERMRTGDRPL TMLSSMTALY 480
481 DGITPEDELI IEDNLGIIQQ ILPINLTSEA VLERGSFPGF KFLEAKPGDG PDGVEGSEDS 540
541 AAPLGPQRRP SSRSMPTCAP PGPAAGNAQN STAENGAETD GVASASDPPP TAAPGAPPRR 600
601 RKRNFFSCIF RSRTDSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
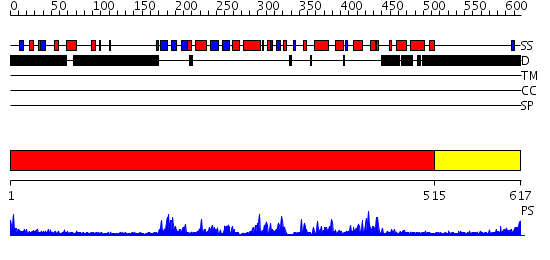
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..514] | 59.09691 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [515..617] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)