













| General Information: |
|
| Name(s) found: |
CE27373
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 549 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
electron transport
[IEA glutamate biosynthetic process [IEA removal of superoxide radicals [IEA glutathione metabolic process [IEA detoxification of mercury ion [IEA |
| Molecular Function: |
disulfide oxidoreductase activity
[IEA NADP or NADPH binding [IEA oxidoreductase activity [IEA FAD binding [IEA oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor [IEA NAD or NADH binding [IEA mercury ion binding [IEA mercury (II) reductase activity [IEA thioredoxin-disulfide reductase activity [IEA glutathione-disulfide reductase activity [IEA oxidoreductase activity, acting on NADH or NADPH [IEA dihydrolipoyl dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALANWLTPV EPECDNDDSP AVEEILGRIE DVPPGTKKMF EVRDRKVLVI NDEGRIYAIN 60
61 GLCSHYNFSL ENGTYAKGRI RCPLHGACFN VRSGDIEDYP GFDSLHSYVV TVNDGNLIIK 120
121 TTEKKLGSDR RIRHLPKMKQ CNDRPVVIIG GGVATATFIE HSRLNGLITP ILVISEESLP 180
181 PYDRVLLSKK PAATGEDIRL RKDDAFYEER NVKFLLKTSV IAVNHKSREV SLSNGETVVY 240
241 SKLIIATGGN VRKLQVPGSD LKNICYLRKV EEANIISNLH PGKHVVCVGS SFIGMEVASA 300
301 LAEKAASVTV ISNTPEPLPV FGSDIGKGIR LKFEEKGVKF ELAANVVALR GNDQGEVSKV 360
361 ILENGKELDV DLLVCGIGVT PATKFLEGSG IKLDNRGFIE VDEKFRTNIS YIFAMGDVVT 420
421 APLPLWDIDS INIQHFQTAQ AHGQHLGYTI VGKPQPGPIV PYFWTLFFFA FGLKFSGCNQ 480
481 GSTKEYTNGD PETGTFIRYF LKKDKVVAVA AGGPSSVASQ FAEIFKKGIE VTLKDLKNSS 540
541 DHTWGHLLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
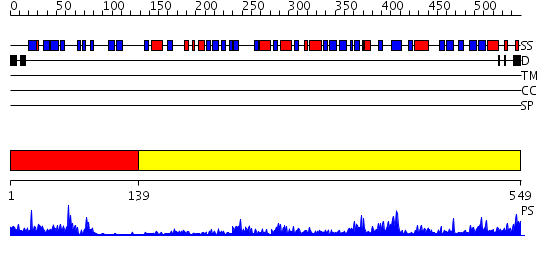
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 15.0 | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | |
| 2 | View Details | [139..549] | 60.69897 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)