













| General Information: |
|
| Name(s) found: |
fbl-1 /
CE26701
[WormBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extracellular region
[IEA proteinaceous extracellular matrix [IC |
| Biological Process: |
extracellular matrix organization
[ISS blood coagulation [IEA |
| Molecular Function: |
calcium ion binding
[IEA serine-type endopeptidase activity [IEA extracellular matrix structural constituent [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRICFLLLAF LVAETFANEL TRCCAGGTRH FKNSNTCSSI KSEGTSMTCQ RAASICCLRS 60
61 LLDNACDSGT DIAKEEESCP SNINILGGGL KKECCDCCLL AKDLLNRNEP CVAPVGFSAG 120
121 CLRSFNKCCN GDIEITHASE IITGRPLNDP HVLHLGDRCA SSHCEHLCHD RGGEKVECSC 180
181 RSGFDLAPDG MACVDIDECA TLMDDCLESQ RCLNTPGSFK CIRTLSCGTG YAMDSETERC 240
241 RDVDECNLGS HDCGPLYQCR NTQGSYRCDA KKCGDGELQN PMTGECTSIT CPNGYYPKNG 300
301 MCNDIDECVT GHNCGAGEEC VNTPGSFRCQ QKGNLCAHGY EVNGATGFCE DVNECQQGVC 360
361 GSMECINLPG TYKCKCGPGY EFNDAKKRCE DVDECIKFAG HVCDLSAECI NTIGSFECKC 420
421 KPGFQLASDG RRCEDVNECT TGIAACEQKC VNIPGSYQCI CDRGFALGPD GTKCEDIDEC 480
481 SIWAGSGNDL CMGGCINTKG SYLCQCPPGY KIQPDGRTCV DVDECAMGEC AGSDKVCVNT 540
541 LGSFKCHSID CPTNYIHDSL NKNRCNRQPS ACGLPEECSK VPLFLTYQFI SLARAVPISS 600
601 HRPAITLFKV SAPNHADTEV NFELQLKTTI VGAPNVLPAI RANFLLQKGE KRNSAVVTLR 660
661 DSLDGPQTVK LQLLLRMSKK GKNFNTYAAN LIVDVAAHKR HNTVHPPLMK IR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
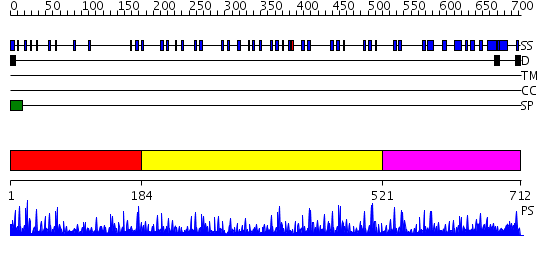
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 4.30103 | No description for 2dtgE was found. | |
| 2 | View Details | [184..520] | 16.522879 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor | |
| 3 | View Details | [521..712] | 2.21 | Domain G2 of nidogen-1; EGF-like domain of nidogen-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|