













| General Information: |
|
| Name(s) found: |
hsf-1 /
CE22380
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
dauer larval development
[IGI locomotory behavior [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP determination of adult lifespan [IGI positive regulation of growth rate [IMP regulation of transcription, DNA-dependent [IEA gamete generation [IMP growth [IMP |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQPTGNQIQQ NQQQQQQLIM RVPKQEVSVS GAARRYVQQA PPNRPPRQNH QNGAIGGKKS 60
61 SVTIQEVPNN AYLETLNKSG NNKVDDDKLP VFLIKLWNIV EDPNLQSIVH WDDSGASFHI 120
121 SDPYLFGRNV LPHFFKHNNM NSMVRQLNMY GFRKMTPLSQ GGLTRTESDQ DHLEFSHPCF 180
181 VQGRPELLSQ IKRKQSARTV EDKQVNEQTQ QNLEVVMAEM RAMREKAKNM EDKMNKLTKE 240
241 NRDMWTQMGS MRQQHARQQQ YFKKLLHFLV SVMQPGLSKR VAKRGVLEID FCAANGTAGP 300
301 NSKRARMNSE EGPYKDVCDL LESLQRETQE PFSRRFTNNE GPLISEVTDE FGNSPVGRGS 360
361 AQDLFGDTFG AQSSRYSDGG ATSSREQSPH PIISQPQSNS AGAHGANEQK PDDMYMGSGP 420
421 LTHENIHRGI SALKRDYQGA SPASGGPSTS SSAPSGAGAG ARMAQKRAAP YKNATRQMAQ 480
481 PQQDYSGGFV NNYSGFMPSD PSMIPYQPSH QYLQPHQKLM AIEDQHHPTT STSSTNADPH 540
541 QNLYSPTLGL SPSFDRQLSQ ELQEYFTGTD TSLESFRDLV SNHNWDDFGN NVPLDDDEEG 600
601 SEDPLRQLAL ENAPETSNYD GAEDLLFDNE QQYPENGFDV PDPNYLPLAD EEIFPHSPAL 660
661 RTPSPSDPNL V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
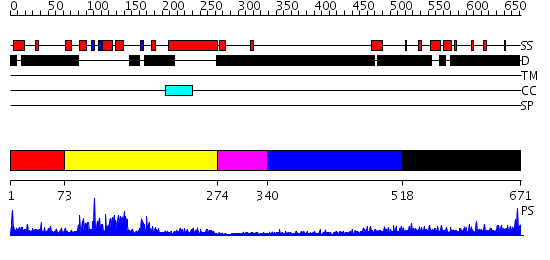
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [73..273] | 43.69897 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 3 | View Details | [274..339] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [340..517] | 2.244997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [518..671] | 5.188991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)