













| General Information: |
|
| Name(s) found: |
rec-8 /
CE21236
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 781 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IDA synaptonemal complex [IDA condensed nuclear chromosome [IDA |
| Biological Process: |
reciprocal meiotic recombination
[TAS embryonic development ending in birth or egg hatching [IMP synaptonemal complex assembly [IDA chromosome segregation [IMP meiotic chromosome movement towards spindle pole [IMP meiosis [IMP double-strand break repair [IDA |
| Molecular Function: |
damaged DNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVSAEVIRK DAVFHVAWIL GTGDSKKLSR REILDQNLPE LCHSIIEMVP ERHRGSATKT 60
61 GLYLLSLLTY GTVLIHQVQV DFLKRDVEKL KELMKKKSFI LLMAERFDRN QELQRKEDKF 120
121 ARLRSKPIMC VEELDRVDLA HLQAIGDELG INGNPGDFIM MDALPNMNQW IDNNSELNAI 180
181 YGCVEPYLRE KEITMHSTFV EGNGSNEHNK ERRNDAVIAD FSQLLFPEIP EITLGEKFPI 240
241 DVDSRKRSAI LQEEQEEALQ LPKEASEIVQ EEPTKFVSIA LLPSETVEQP APQEPIQEPI 300
301 QPIIEEPAPQ LELPQPELPP QLDAIDLVTI PASQQDMVVE YLQLINDLPD DENSRLPPLP 360
361 KDLELFEDVI LPPPAKKSKV EEEEDALERA RRRPSSRPVT PINQTDLTDL HSTVRPEDPS 420
421 FAIDSQIHDV LPQRKKSKRN LPIIHSDDLE IDEAVQKVLQ ADYSSLVRKK EDVIAKIPPK 480
481 TDAVAVLMNL PEPVFSIGYR LPPEVRDMFK ACYNQAVGSP VSDDEEDEDE EEEEEYKYAK 540
541 VCLLSPNRIV EDTLLLEEQP RQPEEFPSTD NINPPRQLQE NPVFENLEYE APPHPIRTAR 600
601 TPTPIKDLKY SVISLFPTPE KRRETSIIAE LNLDPIPVEE IDPLLTMRTE EELENVRRRQ 660
661 KSSLGVQFMR TDDLEEDTRR NRLFEDEERT RDAREDELFF YSSGSLLPNN RLNIHKELLN 720
721 EAEARYPEWV NFNEFTADHD RKKAATAFEG LLLSLKNMKV EAKQEDPYFP ILVRHISHEE 780
781 M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
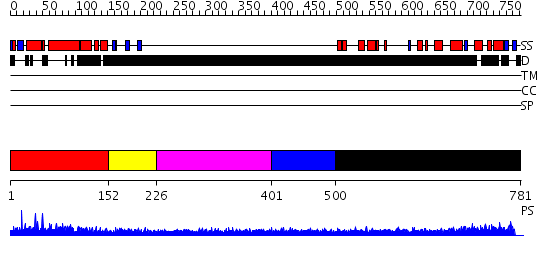
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 50.124939 | No description for PF04825.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [152..225] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [226..400] | 2.234995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [401..499] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [500..781] | 3.34 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)