













| General Information: |
|
| Name(s) found: |
mys-1 /
CE21225
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 458 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA chromatin [IEA |
| Biological Process: |
negative regulation of vulval development
[IMP morphogenesis of an epithelium [IMP nematode larval development [IMP positive regulation of growth rate [IMP post-embryonic body morphogenesis [IMP embryonic development ending in birth or egg hatching [IMP chromatin assembly or disassembly [IEA gamete generation [IMP |
| Molecular Function: |
chromatin binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEPKKEIIE DENHGISKKI PTDPRQYEKV TEGCRLLVMM ASQEEERWAE VISRCRAANG 60
61 SIKFYVHYID CNRRLDEWVQ SDRLNLASCE LPKKGGKKGA HLREENRDSN ENEGKKSGRK 120
121 RKIPLLPMDD LKAESVDPLQ AISTMTSGST PSLRGSMSMV GHSEDAMTRI RNVECIELGR 180
181 SRIQPWYFAP YPQQLTSLDC IYICEFCLKY LKSKTCLKRH MEKCAMCHPP GNQIYSHDKL 240
241 SFFEIDGRKN KSYAQNLCLL AKLFLDHKTL YYDTDPFLFY VLTEEDEKGH HIVGYFSKEK 300
301 ESAEEYNVAC ILVLPPFQKK GYGSLLIEFS YELSKIEQKT GSPEKPLSDL GLLSYRSYWS 360
361 MAIMKELFAF KRRHPGEDIT VQDISQSTSI KREDVVSTLQ QLDLYKYYKG SYIIVISDEK 420
421 RQVYEKRIEA AKKKTRINPA ALQWRPKEYG KKRAQIMF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
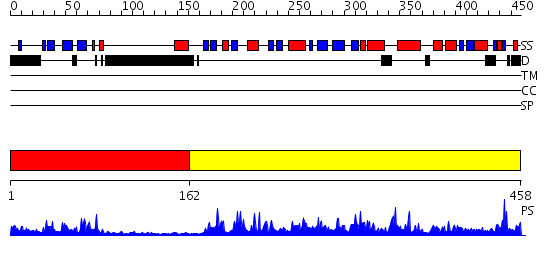
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 15.39794 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [162..458] | 107.0 | No description for 2ou2A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)