













| General Information: |
|
| Name(s) found: |
CE20820
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 336 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
malate metabolic process
[IEA cellular carbohydrate metabolic process [IEA glycolysis [IEA tricarboxylic acid cycle intermediate metabolic process [IEA |
| Molecular Function: |
L-malate dehydrogenase activity
[IEA malate dehydrogenase activity [IEA oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [IEA oxidoreductase activity [IEA malate dehydrogenase (NADP+) activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAPLRVLVT GAAGQIGYSI VIRIADGTVF GKEQPVELVL LDVPQCSNIL EGVVFELQDC 60
61 ALPTLFSVVA VTDEKSAFTG IDYAFLVGAM PRREGMERKD LLAANVKIFK SQGKALAEYA 120
121 KPTTKVIVVG NPANTNAFIA AKYAAGKIPA KNFSAMTRLD HNRALAQLAL KTGTTIGNVK 180
181 NVIIWGNHSG TQFPDVTHAT VNKNGTETDA YAAVGDNAFL QGPFIATVQK RGGVIIEKRK 240
241 LSSAMSAAKA ACDHIHDWHF GTKAGQFVSM AVPSDGSYGI PQGLIFSFPV TIEGGEWKIV 300
301 QGLSFDDFAK GKIAATTKEL EEERDDALKA CDDANI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
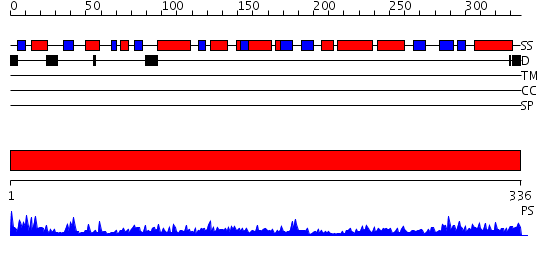
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..336] | 85.522879 | Malate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)