













| General Information: |
|
| Name(s) found: |
ahr-1 /
CE20561
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 602 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA intracellular [TAS |
| Biological Process: |
signal transduction
[IEA regulation of transcription, DNA-dependent [IEA response to xenobiotic stimulus [IDA regulation of transcription [IEA intracellular receptor mediated signaling pathway [TAS |
| Molecular Function: |
DNA binding
[IDA transcription regulator activity [IEA signal transducer activity [IEA aryl hydrocarbon receptor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYASKRRQRN FKRVRDPPKQ LTNTNPSKRH RERLNGELET VAMLLPYDSS TISRLDKLSV 60
61 LRLAVSFLQC KAHFQACLHN SQFLSAGFPM STHSYSYQPH PPIPFSNKVP TIFDLRIGTP 120
121 MLDPEESNFE EISLKSLGGF ILVLNDNGEI YYASENVENY LGFHQSDVLH QPVYDLIHSE 180
181 DRDDIRQQLD SNFHIPTSSA SNQFDVFAPQ NSKYLERNVN ARFRCLLDNT CGFLRIDMRG 240
241 KLMSLHGLPS SYVMGRTASG PVLGMICVCT PFVPPSTSDL ASEDMILKTK HQLDGALVSM 300
301 DQKVYEMLEI DETDLPMPLY NLVHVEDAVC MAEAHKEAIK NGSSGLLVYR LVSTKTRRTY 360
361 FVQSSCRMFY KNSKPESIGL THRLLNEVEG TMLLEKRSTL KAKLLSFDDS FLQSPRNLQS 420
421 TAALPLPSVL KDDQDCLEPS TSNSLFPSVP VPTPTTTKAN RRRKENSHEI VPTIPSIPIP 480
481 THFDMQMFDP SWNHGVHPPA WPHDVYHLTQ YPPTYPHPPG TVGYPDVQIA PVDYPGWHPN 540
541 DIHMTQLPHG FTPDAQKLVP PHPQMSHFTE YPTPSTHHDL HHHPLKQDNF HLISEVTNLL 600
601 GT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
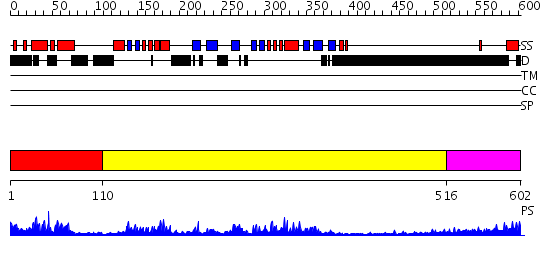
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 8.39794 | SREBP-1a | |
| 2 | View Details | [110..515] | 50.69897 | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | |
| 3 | View Details | [516..602] | 3.248997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)