













| General Information: |
|
| Name(s) found: |
pmr-1 /
CE20426
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA membrane [IEA integral to membrane [IEA Golgi apparatus [IDA |
| Biological Process: |
response to manganese ion
[IMP proton transport [IEA monovalent inorganic cation transport [IEA magnesium ion transport [IEA metabolic process [IEA transport [IEA response to stress [IMP potassium ion transport [IEA response to oxidative stress [IMP cation transport [IEA manganese ion transport [IDA response to calcium ion [IMP calcium ion transport [IDA phospholipid transport [IEA metal ion transport [IEA |
| Molecular Function: |
ATPase activity
[IEA magnesium-importing ATPase activity [IEA manganese-transporting ATPase activity [IDA ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism [IEA monovalent inorganic cation transmembrane transporter activity [IEA hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA calcium ion transmembrane transporter activity [IEA metal ion transmembrane transporter activity [IEA phospholipid-translocating ATPase activity [IEA calcium-transporting ATPase activity [IDA potassium-transporting ATPase activity [IEA magnesium ion binding [IEA ATP binding [IEA copper-exporting ATPase activity [IEA catalytic activity [IEA metal ion binding [IEA calcium ion binding [IEA phosphatase activity [IEA hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIETLTSEQA ASHEVVPCTH QLRTNLEEGL TTAEATRRRQ YHGYNEFDVG EEEPIYKKYL 60
61 EQFQNPLILL LLASAFVSIV MKQYDDAISI TVAVVIVVTV GFVQEYRSEK TLEQLTKLVP 120
121 PTCHVLRDGK EAMMLARELV PGDIVLLNTG DRIPSDLRIA ESFSLQIDES SLTGETEPKH 180
181 KETRSVPAAS ATGSDVEHLT CIAFMGTLVC AGRGRGIVIS TAANSQFGEV VKMMMGEESP 240
241 KTPLQKSMDD LGKQLSIYSF GVIAVIFLIG MFQGRNVVDM FTIGVSLAVA AIPEGLPIVV 300
301 AVTLAIGVMR MAKRRAVVKK MPAVETLGCV TVICSDKTGT LTKNEMTAQA IATPEGKLAE 360
361 ITGIGYSAEG GVVQYQGEQV HQWTHPEFAR IIEAGLVCNN ATIEADKLIG QPTEGAIVVL 420
421 AKKAQLEGVR SEYKRLREMP FSSDTKWMGV QCADAHGQNV YFIKGALDRV LDQCGTYYSS 480
481 DNQRKQCDQY SRQHILEIGK ELGQKGLRVL GLARGESMQS LMFLGMIGMM DPPRPGAADA 540
541 ISIVKASGVD VKLITGDAME TAQSIGQSLG ILSSSDSCLS GQQVDQMSDH DLELVIRQVT 600
601 VFYRASPRHK LKIVKALQAL GEVVAMTGDG VNDAVALKKA DIGVAMGVCG TDVCKEAADM 660
661 ILCDDDFSTM TAAIEEGKAI YHNITNFVRF QLSTKLNSTQ TNYKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
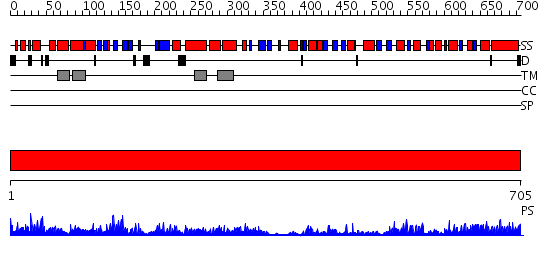
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..705] | 154.0 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M |
| Source: Reynolds et al. 2008. Manuscript submitted |

|