













| General Information: |
|
| Name(s) found: |
daf-19 /
CE17763
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
DNA binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNEEPVPST SSVLSAAEKN VKIETPSRKR KGLDIDALWR KKFKDVENQK SPEKSEKSEK 60
61 TIKEEPLETH PSGERKLRSA SKDSKSLSKE THNTISTRSS SSGTPRKKME PEDVKPNIKM 120
121 LKKSLPVSFQ CSNLNDGTVG DEMEVIQHST DDPNGTREEF DYNQIEYGNA VTPNGTYTLY 180
181 APDSNYAPYY QYSNNMQSSP QDNAPYLVPA PQSPVSIDDA DSINLPNNQR ASPATVNWLF 240
241 ENYEIGEGSL PRCELYDHYK KHCAEHRMDP VNAASFGKLI RSVFHNLKTR RLGTRGNSKY 300
301 HYYGIRLKDS STLHSMQHPQ PQIIQNTYTP PMQLPQRDAY ADTVNQVAAI KYMDVEIPDK 360
361 RARKDNSSSS SSCRDSVSPC MDLPAQQTIQ HTPVHSVIPS TQQPAPLHNN NVVSYVVTES 420
421 DKAAMGKIDL PIVPFPDKDA LMATIGFRKL GVGEEELNSL IDIYEILCRE ILALIKNIDF 480
481 ASVEDTWSKF WSGNFGVDRD HISALCTLDQ VQDYIIEVDL ALYQTIVDTL IPNVLLSELS 540
541 TGMTQTCRTF AKNIDVYLRK SLMLANLGEF FVKKKIQAIK YLQQGLKRYT SLNHLAHASR 600
601 GVLMKPEQVQ QMYQDYIRVD INTVHQQAGW ICGCDSVMVH HVNNAFKHNL QRMSAMEVWA 660
661 EWLESIVDQV LAKYHDKPAN VIANVGKQFL LNWSFYTSMI IRDLTLRSAM SFGSFTLIRL 720
721 LADDYMYYLI ESKIAKAGKQ QLITVIRADK DWPLTTNPQE YIVVANDNDE DLDLEKAGLL 780
781 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
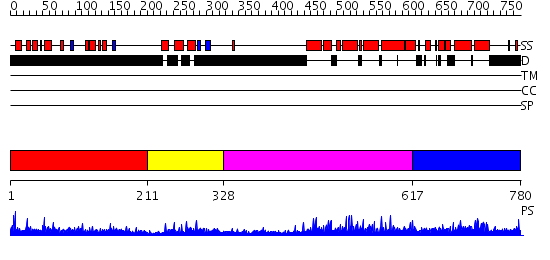
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 3.522879 | Sec24 | |
| 2 | View Details | [211..327] | 20.39794 | Class II MHC transcription factor RFX1 | |
| 3 | View Details | [328..616] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [617..780] | 5.07198 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)