













| General Information: |
|
| Name(s) found: |
nhr-31 /
CE17474
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 583 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQLDVEDWE YWGDEYLEEE PTYAIRPGTR VVKVERVPMM RGDLRTSGAT SSSGPATSYI 60
61 IRPSDKQPTV SSGGSQNGDS VCAVCGDGIA KLHYGVLACY GCKGFFRRTL TGKYRYACRF 120
121 SNNCIVDKFQ RNSCRYCRFQ RCIQAGMDPK AVRPDRDQTG KQKVPRIKKK QIDEELLNHM 180
181 MRLQGDDWSR KLPVETRILL MQLMSIEDKV VKGDNNMSAQ NTAKDPKSIS LREMFESKPA 240
241 LDGRRMEIGY EPFRMARTEE LGVIAHRRAI AAVDWVDSLT EIADAVDTED KVALVKSCYS 300
301 PLTIFNFSAR TAQNTKNPDI LCLCSHSFVP RRLPPEFNET NHLSNFLIDR TLNELVAPLR 360
361 KLNLKEEEIV PLKAIIILNP NAKGLSEHAR HAISELRDKV QDMLFQIVKE LHPIYSASSR 420
421 FGNLLLLLPT ITTLSGLMSE NMHFCQALGG RASGDNLLAE MFGDRTFDDQ LISSSISPPL 480
481 FENPAEICLE SISPPMSDRR FVRRVDVATQ TNDDLLSSGP YLPHSNSCSS MLNAGYSPPM 540
541 LSTSPFPLLD DNDSAFQNDI SLTELNGCEE FFSQLMDQPI IDS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
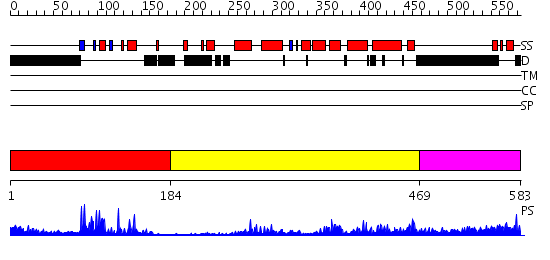
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 29.0 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 2 | View Details | [184..468] | 46.30103 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA | |
| 3 | View Details | [469..583] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)