













| General Information: |
|
| Name(s) found: |
prmt-2 /
CE16546
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
protein amino acid methylation
[IEA |
| Molecular Function: |
protein methyltransferase activity
[IEA methyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFLEKINQKT GEREWVVAEE DYDMAQELAR SRFGDMILDF DRNDKFLAGL KTTIAEKKHE 60
61 NTDGKVHVLD IGTGTGLLSL MAAREGADKV TALEVFKPMG DCARHITSNS PWSDKITVIS 120
121 ERSTDVSQIG GSRADIIVAE VFDTELIGEG ALRTFKEALE RLAKPGCRVV PSTGNVYIVP 180
181 VESHLLKMFN DIPRLNGEKD EEPLGRCSGT AAVFDVQLSE MKTHEFRELS EPIVAFKFDF 240
241 EHEEKIIFDE SFVREAVAHS SGTIDALLMW WDIDMDRNGT TFIDMGPKWK NKNNYALMVQ 300
301 QCWERVCIYS RNTNFYQLLF FSKSRSYCVC GLHSMLSRQT VYHVNEMFEN QKFKDEVDKL 360
361 SKGLHVATVG EGSFLGLLAA KTAKRVTIID GNERFRDIFF KYIHYYKLTN VEIIEKVTSL 420
421 TDSPDIVLAE PFYMSAMNPW NHLRFLYDVE VLKMMHGDEL RVEPHMGVLK AIPEKFEDLQ 480
481 NIASDVGTVN GFDLSFFDEI STKARTATDA IVDEQSLWEY AGIVKGDAVE ILRFPIDGRV 540
541 SSQKCVVNID NMSSSNAIPM WMEWEFGGIN LSTGLLSISS AGVPEWNKGY KQGVYFPITA 600
601 LRNDKSLCLH ALFDKSTGDI NFQFGKSEDS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
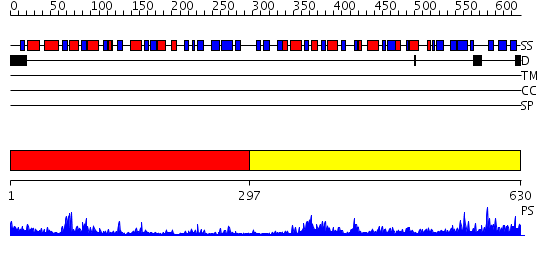
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..296] | 35.0 | CmaA1 | |
| 2 | View Details | [297..630] | 77.522879 | Structure of the Predominant protein arginine methyltransferase PRMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)