













| General Information: |
|
| Name(s) found: |
hsp-1 /
CE09682
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 640 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to heat
[IDA nematode larval development [IMP determination of adult lifespan [IGI embryonic development ending in birth or egg hatching [IMP protein folding [IEA reproduction [IMP growth [IMP iron-sulfur cluster assembly [IEA |
| Molecular Function: |
ATPase activity
[IEA ATP binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKHNAVGID LGTTYSCVGV FMHGKVEIIA NDQGNRTTPS YVAFTDTERL IGDAAKNQVA 60
61 MNPHNTVFDA KRLIGRKFDD PAVQSDMKHW PFKVISAEGA KPKVQVEYKG ENKIFTPEEI 120
121 SSMVLLKMKE TAEAFLGTTV KDAVVTVPAY FNDSQRQATK DAGAIAGLNV LRIINEPTAA 180
181 AIAYGLDKKG HGERNVLIFD LGGGTFDVSI LTIEDGIFEV KSTAGDTHLG GEDFDNRMVN 240
241 HFCAEFKRKH KKDLASNPRA LRRLRTACER AKRTLSSSSQ ASIEIDSLFE GIDFYTNITR 300
301 ARFEELCADL FRSTMDPVEK SLRDAKMDKS QVHDIVLVGG STRIPKVQKL LSDLFSGKEL 360
361 NKSINPDEAV AYGAAVQAAI LSGDKSEAVQ DLLLLDVAPL SLGIETAGGV MTALIKRNTT 420
421 IPTKTAQTFT TYSDNQPGVL IQVYEGERAM TKDNNLLGKF ELSGIPPAPR GVPQIEVTFD 480
481 IDANGILNVS ATDKSTGKQN KITITNDKGR LSKDDIERMV NEAEKYKADD EAQKDRIGAK 540
541 NGLESYAFNL KQTIEDEKLK DKISPEDKKK IEDKCDEILK WLDSNQTAEK EEFEHQQKDL 600
601 EGLANPIISK LYQSAGGAPP GAAPGGAAGG AGGPTIEEVD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
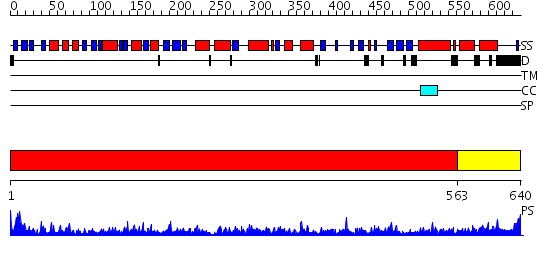
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..562] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [563..640] | 54.221849 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)