













| General Information: |
|
| Name(s) found: |
tkt-1 /
CE09163
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nematode larval development
[IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP terpenoid biosynthetic process [IEA growth [IMP |
| Molecular Function: |
aldehyde-lyase activity
[IEA thiamin pyrophosphate binding [IEA 1-deoxy-D-xylulose-5-phosphate synthase activity [IEA oxidoreductase activity [IEA transketolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVSEQLRND LEDAANRMRI SSIEMTCASK SGHPTSSTSA AEIMSTLFFS EMKYDVAEPK 60
61 SASADRFVLS KGHACPILYA AWEEAGLLSH EQVLSLRKID SDIEGHPTPR LNFIDVATGS 120
121 LGQGLGVATG MAYVGKYIDK ASYRVFCLLG DGESAEGSVW EAAAFASIYK LDNLVAIVDV 180
181 NRLGQSQATS LGHDVETYKA RFAAFGFNAI IVNGHNVDEL LAAYETARST KGKPTAIIAK 240
241 TLKGKGIEGI ENEDNWHGKP VPEGTVNAIK ARFHGSQKGK LVAQKPINDA PAIDLHVGSI 300
301 KMAAPEYKKG DKVATRAAYG TALAKLGDAS PRVIGLDGDT KNSTFSDKLL KKHPDQFIEC 360
361 FIAEQNLVGV AVGAQCRDRT IPFTSTFAAF FTRATDQIRM AAVSFANLKC VGSHVGVSIG 420
421 EDGPSQMALE DLAIFRTIPG ATVFYPTDAV SAERATELAA NTKGVVFIRT GRPALPVLYD 480
481 NEEPFHIGQA KVVKQSAQDK IVLVGSGVTL YESLKAAEEL EKEGIHATVI DPFTIKPLDG 540
541 KTIAEHALKV GGRVVTTEDH YAAGGIGEAV SAALADYPTI RVRSLNVKEV PRSGPPDALV 600
601 DMYGISARHI VAAVKNFQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
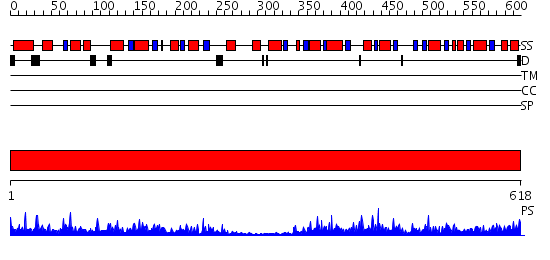
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..618] | 170.0 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)