













| General Information: |
|
| Name(s) found: |
aha-1 /
CE08377
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 453 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA intracellular [TAS |
| Biological Process: |
locomotory behavior
[IMP signal transduction [IEA positive regulation of growth rate [IMP regulation of transcription, DNA-dependent [IEA response to xenobiotic stimulus [IDA regulation of transcription [IEA positive regulation of multicellular organism growth [IMP intracellular receptor mediated signaling pathway [TAS positive regulation of locomotion [IMP |
| Molecular Function: |
DNA binding
[IDA aryl hydrocarbon receptor nuclear translocator activity [ISS transcription regulator activity [IEA signal transducer activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQDIFMDPW QSATSFAMED EDMGMPSGKY ARMEDEMGEN KERFARENHS EIERRRRNKM 60
61 THYINELAEM VPQCASLGRK PDKLTILRMA VSHMKGIRGH TAQDETSYKP SFLTDQELKH 120
121 LILEAANGFL FVVCCQTGKV LYVADSITPV LNLKQEDWLQ RNLNELIHPD DQDKIRDQLC 180
181 GSEVSVNKVL DLKSGSVKRE GASTRVHMSC RRGFICRMRV GALEPLHRLR NRRPLFQHAG 240
241 QNYVVMHCTG YIKNAPPQGI NAPASSCLVA IARLQVASMP VCADPTSTNQ FSVRVSEDGK 300
301 MTFIDARVSD LIGLSSDQLI GRYWWNLAHP ADEKTLQDSF VALLSDQPMR INIRVRTSTD 360
361 YIPCTVSAYK FMNPYSEQFE YVVATHQIAP QEDINNWVTA PTVPQPQASE FGELGGAPSA 420
421 VDYGQSSSGG WRPEAQGAPQ AQWQWDPMNG YNQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
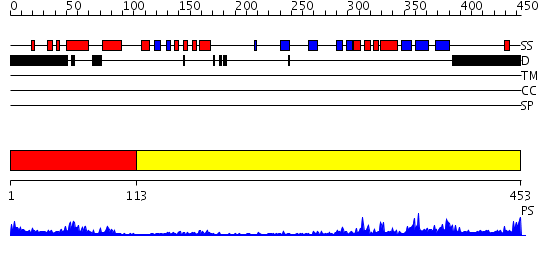
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 5.522879 | SREBP-1a | |
| 2 | View Details | [113..453] | 26.69897 | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)