













| General Information: |
|
| Name(s) found: |
hsp-3 /
CE08177
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 661 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum lumen
[ISS endoplasmic reticulum [IEA |
| Biological Process: |
endoplasmic reticulum unfolded protein response
[IEP nematode larval development [IMP cell morphogenesis [IEA response to chemical stimulus [ISS protein folding [IEA protein secretion [ISS response to stress [ISS growth [IMP positive regulation of multicellular organism growth [IMP iron-sulfur cluster assembly [IEA |
| Molecular Function: |
ATPase activity
[IEA ATP binding [IEA protein binding [ISS calcium ion binding [ISS unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTLFLLGLI ALSAVSVYCE EEEKTEKKET KYGTIIGIDL GTTYSCVGVY KNGRVEIIAN 60
61 DQGNRITPSY VAFSGDQGDR LIGDAAKNQL TINPENTIFD AKRLIGRDYN DKTVQADIKH 120
121 WPFKVIDKSN KPSVEVKVGS DNKQFTPEEV SAMVLVKMKE IAESYLGKEV KNAVVTVPAY 180
181 FNDAQRQATK DAGTIAGLNV VRIINEPTAA AIAYGLDKKD GERNILVFDL GGGTFDVSML 240
241 TIDNGVFEVL ATNGDTHLGG EDFDQRVMEY FIKLYKKKSG KDLRKDKRAV QKLRREVEKA 300
301 KRALSTQHQT KVEIESLFDG EDFSETLTRA KFEELNMDLF RATLKPVQKV LEDSDLKKDD 360
361 VHEIVLVGGS TRIPKVQQLI KEFFNGKEPS RGINPDEAVA YGAAVQGGVI SGEEDTGEIV 420
421 LLDVNPLTMG IETVGGVMTK LIGRNTVIPT KKSQVFSTAA DNQPTVTIQV FEGERPMTKD 480
481 NHQLGKFDLT GLPPAPRGVP QIEVTFEIDV NGILHVTAED KGTGNKNKIT ITNDQNRLSP 540
541 EDIERMINDA EKFAEDDKKV KDKAEARNEL ESYAYNLKNQ IEDKEKLGGK LDEDDKKTIE 600
601 EAVEEAISWL GSNAEASAEE LKEQKKDLES KVQPIVSKLY KDAGAGGEEA PEEGSDDKDE 660
661 L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
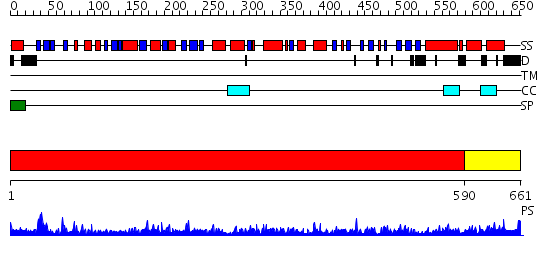
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..589] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [590..661] | 56.0 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|