













| General Information: |
|
| Name(s) found: |
vha-15 /
CE07497
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V1 domain
[IEA peripheral to membrane of membrane fraction [IEA |
| Biological Process: |
collagen and cuticulin-based cuticle attachment to epithelium
[IMP nematode larval development [IMP locomotion [IMP embryonic development ending in birth or egg hatching [IMP ATP synthesis coupled proton transport [IEA growth [IMP |
| Molecular Function: |
ATP binding
[IEA proton-transporting ATPase activity, rotational mechanism [IEA hydrogen ion transporting ATP synthase activity, rotational mechanism [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEVPHHNIP AVDMLNATSR LQLEAQELRN NKPNWGSYFR SQMIQEDDYN FITSFENAKS 60
61 KEERDQVLAA NNANGQAAKT MANLITQVAK DQNVRYVLTL FDDMLQEDKS RVELFHSAAA 120
121 RQKRTVWSQY LGILQRQDNF IVNQMSSIIA KLACFGTTRM EGQDLQYYFS FLKEQLKNST 180
181 TNDYMNTTAR CLQMMLRHDE YRHEFVDSDG VQTLVTALNG KTNFQLQYQL IFAVWCLTFN 240
241 ADIARKAPSL GLIQALGDIL SESTKEKVIR IILASFVNIL SKVDEREVKR EAALQMVQCK 300
301 TLKTLELMDA KKYDDPDLED DVKFLTEELT LSVHDLSSYD EYYSEVRSGR LQWSPVHKSE 360
361 KFWRENASKF NDKQFEVVKI LIKLLESSHD PLILCVASHD IGEYVRHYPR GKTVVEQYQG 420
421 KAAVMRLLTA EDPNVRYHAL LAVQKLMVHN WEYLGKQLDS DVQTDTVAVK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
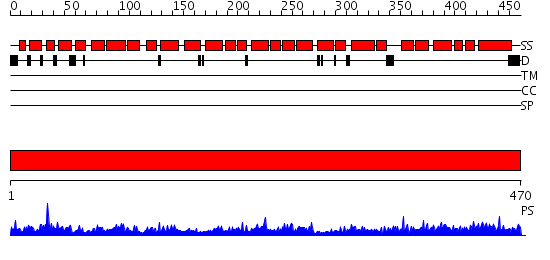
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..470] | 78.39794 | Regulatory subunit H of the V-type ATPase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)