













| General Information: |
|
| Name(s) found: |
plc-4 /
CE07421
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 751 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA lipid metabolic process [IEA |
| Molecular Function: |
phosphoinositide phospholipase C activity
[IEA phospholipase C activity [IEA calcium ion binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKSVSPEEQ AAAQAADEEK TKEELEWAEK GSLLCRIKNQ KVKEMALVTI KSKQFLNYYS 60
61 SYWFNFVPNA LKSVALSELL EVRSGYQTDN LQRASKKYEF QELAPESRCF SVIFSHAKFL 120
121 HKSVDFSADS KETRDKWVSV LTHLISVAKH QRVVFNETAW LIDKFQQADT NKNGLLSFDE 180
181 VWNLLKRMNL QISERYAKAI FRESEFENSR DNKLNEKEFL NFFERLTDRP DLRFVMTQAS 240
241 SDNVETLTVA DLQRFLTEEQ GFENVDLKKA EQILTTFEQT VQDKQKEKLM GLMGMRRLMQ 300
301 SRWGNVFKPG HESIFQDMDQ PLTHYFVNSS HNTYLTGLQV KGEATVEGYI SALRKGARLL 360
361 ELDLFDGEHG EPVITHKRTF IESITLRNSL EAIKRTAFET SPYPVILTLE NHVGFVQQAV 420
421 MADLFKEILG DSLYIPPKDS HRHPLSSPNK LKRKFLLRGK KIILEEDIEE PDEDDSPTDK 480
481 DKHHVHPHPV APELSALIGL PSVKLSHNIY QDVNKHPFDG SPSLSENKVY TMFEAAVPIF 540
541 TYTAERLVKS YPKGLRQDSS NMHPMVSWLC GIQSVAMNFQ TAGEELDLNA GLFRINGNCG 600
601 YVLKPSCLLD GVDPRSMAKP KLKLGIGLFS AQYLPKSEPG KEIIDPYVSV QIFGIPRDET 660
661 KAKTRIIKDN GFNPEWRDNF YFTLSCPELA IIRFCVKDFD STSSNDFVGE FSIPVMSLRT 720
721 GFSQIQLNTG YQHTLDPSAS LFVRIAMEEE Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
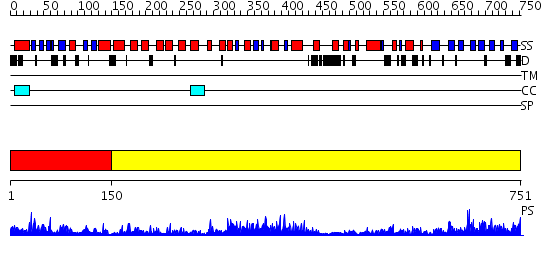
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 23.0 | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | |
| 2 | View Details | [150..751] | 152.0 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)