













| General Information: |
|
| Name(s) found: |
CE06744
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 631 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
carbohydrate metabolic process
[IEA locomotory behavior [IMP nematode larval development [IMP nucleotide-sugar metabolic process [IEA extracellular polysaccharide biosynthetic process [IEA GDP-mannose metabolic process [IEA oviposition [IMP steroid biosynthetic process [IEA biosynthetic process [IEA galactose metabolic process [IEA cellular metabolic process [IEA growth [IMP |
| Molecular Function: |
3-beta-hydroxy-delta5-steroid dehydrogenase activity
[IEA NADP or NADPH binding [IEA coenzyme binding [IEA catalytic activity [IEA UDP-glucose 4-epimerase activity [IEA steroid delta-isomerase activity [IEA dTDP-glucose 4,6-dehydratase activity [IEA L-aminoadipate-semialdehyde dehydrogenase activity [IEA ADP-glyceromanno-heptose 6-epimerase activity [IEA dTDP-4-dehydrorhamnose reductase activity [IEA GDP-mannose 4,6-dehydratase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVYTPKNVVI TGGCGFIGSN FVNYIHDAWP TCNFVNIDKL ILNSDTQNVA ESVRNSPRYK 60
61 LVLTDIKNEA AILNVFEQNE IDTVIHFAAD CTSTRCYNET AEAVQNNVLS FIQFLETVRT 120
121 YGKIKRFVHI STDEVYGDSD LSENEQGKVE FSRLVPGNPY AATKIAGEAY VRAYQTQYNL 180
181 PIVTARMNNI YGPNQWDVKV VPRFIEIAKV RGEYTIQGSG KQLRSWLFVD DASAGLKAVC 240
241 EKGELHEIYN LGTYYEKNVA DLAKTIQEEV DLQLGRAHEP PKYKSIPDRP YNDLRYLISI 300
301 EKAKNDLGWE PTTSFDDGMR HTVASALKEH KHVKMHVAIY GGKGYVGQEL QHVLNDRHIP 360
361 YVLATKKVGF DSDEEVEREL ALLGVTHVIC VTGRTHGPGC NTIEYLEGRA DKVFINVRDN 420
421 MYSATILAHM CRKLGLHYTY IGTGYMFAYD KEHPIGGAEF KEEDEPTFFG SAYSVVKGFT 480
481 DRQMNYFNQN GWENLNVRIT LPLSLDLEQP RNLLSKIIKY KELFDIPVSL TILPDCMNAM 540
541 CNLMEQRSGG TLNLVNPEPI SLYEVVKIYK EIVDETVNPT SIGVETERAQ HLLATKGNCA 600
601 LDTEKLQSLA PVLSAKQSLI KHFSEMTSAA K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
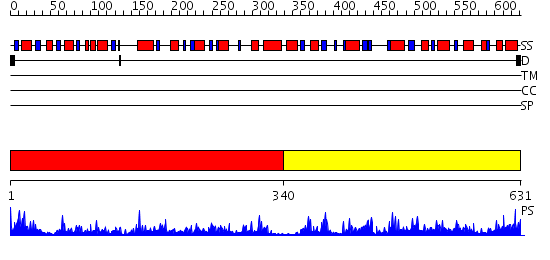
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..339] | 92.09691 | dTDP-glucose 4,6-dehydratase (RmlB) | |
| 2 | View Details | [340..631] | 41.0 | GDP-4-keto-6-deoxy-d-mannose epimerase/reductase (GDP-fucose synthetase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)