













| General Information: |
|
| Name(s) found: |
CE05666
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
hermaphrodite genitalia development
[IMP uracil salvage [IEA biosynthetic process [IEA reproduction [IMP |
| Molecular Function: |
nucleotide binding
[IEA ATP binding [IEA uracil phosphoribosyltransferase activity [IEA uridine kinase activity [IEA nucleoside-triphosphatase activity [IEA kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNQSGGDSE DSTSPRAAGC RTRRRTMSGG RAEHHLLTTK TGKKIYTKGR PPWYDKKGKS 60
61 LKHPFVIGVC GGSASGKTTV AEKIVERLGI PWVTILSMDS FYKVLTPEEI KAAHESRYNF 120
121 DGPNAFDFDL LYEVLKRLRE GKSVDVPVYD FNTHSRDPNS KMMYGADVLI FEGILAFHDE 180
181 RIKNLMDMKV FVDTDGDLRL ARRIVRDVTD RGRDIDGIME QYFTFVKPAF DKYIAPCMDS 240
241 ADLIVPRGGE NDVAIDMIVQ NVMAQLVERG YDRNQNNRDR HDLVRDDLPD CLPENLFILK 300
301 ETPQVKGLVT FVRDRETSRD NHIFYSDRLM RILIEECMNH MPYKDVEIEM AGGRKTIGKR 360
361 KDAQICGLPI MRAGECMETA LRSIVKDCVI GKILIQTNET TFDPELHYIR LPPHITRYKV 420
421 IIMDATVTTG SAAMMAIRVL LDHDVKEEDI FVASLLMGQQ GAHALAYAFP KVKLITTAMD 480
481 HQMTENCYLI PGMGNFGDRY YGTGLEQDMD EPFDM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
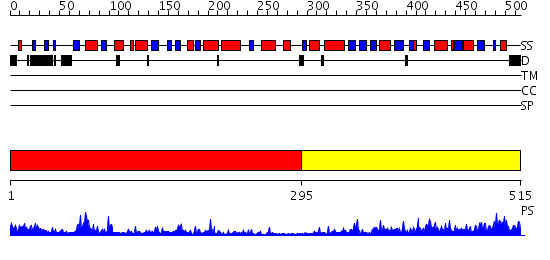
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..294] | 45.221849 | Crystal structure of human uridine-cytidine kinase 2 complexed with a feedback-inhibitor, CTP | |
| 2 | View Details | [295..515] | 54.39794 | Uracil PRTase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)