













| General Information: |
|
| Name(s) found: |
cel-1 /
CE05197
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 573 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
DNA repair
[IEA molting cycle, protein-based cuticle [IMP mRNA capping [IEA DNA replication [IEA post-embryonic body morphogenesis [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP protein amino acid dephosphorylation [IEA DNA recombination [IEA gamete generation [IMP mRNA processing [IEA positive regulation of multicellular organism growth [IMP growth [IMP reproduction [IMP positive regulation of locomotion [IMP locomotory behavior [IMP nematode larval development [IMP morphogenesis of an epithelium [IMP dephosphorylation [IEA |
| Molecular Function: |
ATP binding
[IEA DNA ligase (ATP) activity [IEA protein tyrosine phosphatase activity [IEA RNA-dependent ATPase activity [ISS phosphatase activity [IEA mRNA guanylyltransferase activity [IEA protein tyrosine/serine/threonine phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLPDRWLHC PKTGTLINNL FFPFKTPLCK MYDNQIAERR YQFHPAEVFS HPHLHGKKIG 60
61 LWIDLTNTDR YYFREEVTEH ECIYHKMKMA GRGVSPTQED TDNFIKLVQE FHKKYPDRVV 120
121 GVHCTHGFNR TGFLIAAYLF QVEEYGLDAA IGEFAENRQK GIYKQDYIDD LFARYDPTED 180
181 DKILAPEKPD WEREMSIGMS TQIDNGRPST SQQIPATNGN NNQNGNQLSG GGDNSKLFMD 240
241 GLIRGVKVCE DEGKKSMLQA KIKNLCKYNK QGFPGLQPVS LSRGNINLLE QESYMVSWKA 300
301 DGMRYIIYIN DGDVYAFDRD NEVFEIENLD FVTKNGAPLM ETLVDTEVII DKVEINGAMC 360
361 DQPRMLIYDI MRFNSVNVMK EPFYKRFEII KTEIIDMRTA AFKTGRLKHE NQIMSVRRKD 420
421 FYDLEATAKL FGPKFVQHVG HEIDGLIFQP KKTKYETGRC DKVLKWKPPS HNSVDFLLKV 480
481 EKKCKEGMLP EWIGYLFVQN LSDPFGTMKA TATLKKYHNK IIECTLLVDN QGRPKEWKFM 540
541 RERTDKSLPN GLRTAENVVE TMVNPVTETY LIE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
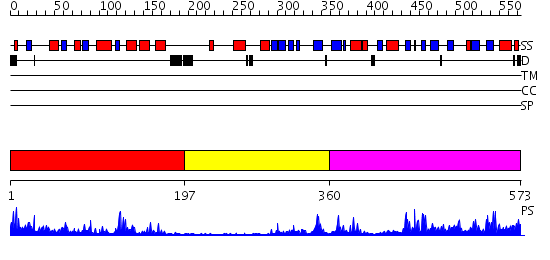
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 32.69897 | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | |
| 2 | View Details | [197..359] | 2.38 | RNA guanylyltransferase (mRNA capping enzyme); RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain | |
| 3 | View Details | [360..573] | 53.522879 | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)