













| General Information: |
|
| Name(s) found: |
brf-1 /
CE02738
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 759 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
nematode larval development
[IMP positive regulation of transcription [IEA gamete generation [IMP regulation of transcription [IEA growth [IMP |
| Molecular Function: |
zinc ion binding
[IEA transcription activator activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRTCSNCGS SEIDEDAARG DATCTACGTV LEESIVVTEN QFQERAGGSG HTLVGQFVSS 60
61 ERAAANNFNG MGSQESREMT YAKGRKVIDE LGSQLRINSH CMNTAFNFYK MCVSRNLTRG 120
121 RNRSSVVAVC MYITCRLENT AHLLLDFSDV TQINVFDLGR NLNYLSRSLR INLPSTDPCL 180
181 YIMRFACVLD FGDKQKEVVN LATRLVQRMK RDWMSTGRRP TGICGAALLI AARSLNFNRS 240
241 INDIVRVVHI SEGVIRKRLD EFSQTPSGSL TIDEFSTVDL EHSEDPPAYR ESRRKAREDQ 300
301 LRKEAEQAES MKEQLGEMEA HVEAALDKKR KEKFSKSPYA RMISENLGKE NDKMQTLTET 360
361 VVESSGLEKG ADEMVRNEII NTVFNAAEED PCTSNSLEKY DKYRPSLESL GIKRTSEPEP 420
421 EPVPHPIVNA DLEEDISDSE IDSYILTESE VAIKTDYWMK ANGEAMKEIE EKKRERELNG 480
481 GVKKKKPRSN RKTDTTSTSV ASAVEKVIAE KKLSNKVNYE MLKDLETMAT GIKRDIREST 540
541 PAPVTPISIK VDSSDVTPDT LSKSEKIARG KNIRSQASES TIQKLRSIFD LTEECSETSK 600
601 NSSPKVNLKV ESASPSTSEV SSIEHKPFVP PARSRYAKVK PIIGAKKLAA LNEVKNVHTV 660
661 PEPSAPAEPI VSEAPLQVKT SSSDPIVTST EVPSVAEPPA PIQNVEEAKP IVPVKSRYAK 720
721 AKPNLKPIKQ KTAVDVVTVG EGESSEPSAK RTKAEHLES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
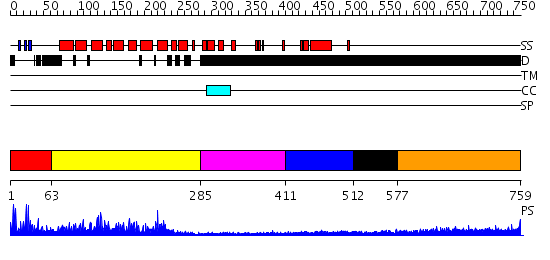
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | 1.090992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [63..284] | 64.0 | Transcription factor IIB (TFIIB), core domain | |
| 3 | View Details | [285..410] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [411..511] | 2.58 | TBP-binding domain of the transcription factor IIIb Brf1 subunit | |
| 5 | View Details | [512..576] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [577..759] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)