













| General Information: |
|
| Name(s) found: |
rpn-3 /
CE00101
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome complex
[IEA |
| Biological Process: |
embryonic cleavage
[IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP reproduction [IMP growth [IMP |
| Molecular Function: |
enzyme regulator activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPKTGETVV EKMEVDEAKQ DVPATEPAKD LNAIAVENIK EQLAALDKGE EHLITRVLQV 60
61 LPKTRKQIND NVLYKLVSSH LSSDAQFAEG MLKYLHYTPA ADTEVQPMDT SNVKTKSPKK 120
121 GVKPVFASPE SDCYLRLLVL LHLYAQKKNT EALALGENQL TSIYNFDRRT LDGLAAKTLY 180
181 FLCVIYEREG RLFDHQGFLN SRLRTATLRN FSESQAVLIC WLLRCYLINR QYQSAAHLVS 240
241 KVAFPDNASN NDLARYMYYQ GRIKALQLDY NSAAGYFLQA QRKAPQEGAI GFKQAVQKWV 300
301 VVIGLLQGEI PDRSVFRQPI YRKCLAHYLD LSRGVRDGDV ARFNHNLEQF KTQFEADDTL 360
361 TLIVRLRQNV IKTAIKQISL AYSRIYIKDI AKKLYITNET ETEYIVAKAI ADGAIDAVIT 420
421 SDVRDGPRYM QSSETADIYR TSEPQAHFDT RIRYCLELHN QAVKALRYPP KKKIAVETIE 480
481 QAREREQQEL EFAKELADED DDDF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
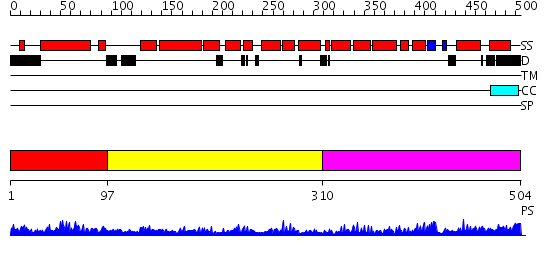
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 1.070995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [97..309] | 7.0 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 3 | View Details | [310..504] | 7.522879 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)