













| General Information: |
|
| Name(s) found: |
rav1 /
SPBC1105.10
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1297 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RAVE complex
[IPI nucleus [IDA cytoplasm [IDA cytosol [IDA endosome membrane [IDA |
| Biological Process: |
regulation of proton-transporting ATPase activity, rotational mechanism
[TAS vacuolar proton-transporting V-type ATPase complex assembly [TAS response to zinc ion [IMP response to calcium ion [IMP response to drug [IMP late endosome to vacuole transport [TAS vacuolar acidification [IMP |
| Molecular Function: |
protein complex binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLAFTNPGT PLKRTAAYDN VQWNQNRILT FGTRYTVLII VNNYESYQNI QFGESIGAVA 60
61 LDQRTGFLAI AHGQVLDIYK PADSLKWTHH YNYITQLSED ITQLSWGYNF DLLLCASSYT 120
121 KLINFSEDKP RLVWSQNRIS DSFLSSTISS DACFIASIET KNPLAKVWQR TSPQGTVSSC 180
181 DDFSYLVHPC RVRFSQWSKH LHPDASGSTP FLSTVGYDNV LRIWQVGSRF GSQLMHLSCF 240
241 LNLRKYYKSD QPPYCCFIDK DDFTIALAHA ISRHSGQVHK DRILNRLLAL ASGQPHIFLL 300
301 FTFECLYIFS LVIDQETLSS FELINQVPTN FPKGALQPDG FIPIYRTSKH EFYDYNIILF 360
361 FQNSAREYMF SLTDFFYSDF ELRVSYDFYG HDYPIKSLTR TSNGHGIVSR DVNRICMFQS 420
421 YISQDGKNQL ICRFRLLLGE SDFILPLYAG EYAAVLNLNS LKLWHCNEAI PPTLLCTCRQ 480
481 VPTSPIITFF ILPVQNNDNS ALCALTEGGH AWFWSVSNEA HDGSAVLRFI DRVNFTKNLN 540
541 IASAVDVMGW SSTLNLSSLS SFDQEVFQSI SKDGLLQTWM ARVLGDDKAE ISEISKVQTS 600
601 IKSATMIKGS TSKKVAVVSE NNRRLSIFDT RSSEFSEKEE FSSVFDDYGP INDLDWTSTP 660
661 NSHSILAVGF LHDVILLCQN RRSYMNDIPC WSRIRRYNLL EYTNSLISDS SWLDNGTLVT 720
721 AIGNGLFYFD NSLPEDQSLL FPASLKRSAK NIFDLAYQLN GILPVFHPQL LQQLLLRNQP 780
781 FLFTNILLRF YLCLKDDVDM HFLLNMDCSE FYCCDEEISK DLLIKSLSQN SAEILNPAEL 840
841 DFEETSVSKH LGLYIDEMIK KLTALLKVKK VKTLTRSSQF LLLNLIEAFN KARALRDSLD 900
901 LNGKRYCIML NQYVLSKHQR QLTSLPTREM AWAFHSSNKA VLLDHTKKLH GKPLLWNAVE 960
961 EYAIPFWLNE QLLKDVFLEL SRNHYAEIDD RNPENVSLYH IALHKINVFR DLWRLASWHK1020
1021 ESARTVKLLS NDFSKKKWKV VASKNAFALL SKHRYFYASA FFLLADSCYD AAKVCIRNLK1080
1081 SISLAIAMTR VYEGDDGPTL KRLINEYVLP LAVSQNDRWL TNWSFTILKQ EKKALQSLVS1140
1141 PIRSLVSDND FIQLYTSDKN DQEIHENKDS QRNSTNEPVR NKFPSDDPAF ILLYECLRSS1200
1201 NVQIDESLEY RFVLHNANVY CQLGCDLIAL GLLRNWKFQK NPATAHLETE DYMVTDKQSA1260
1261 VEKTHDSPKL GKTLMGQDIR PTPDDVPEFN ESAFSFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
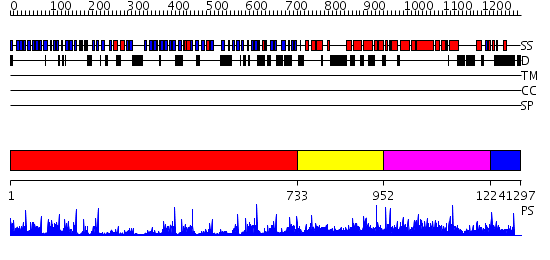
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..732] | 27.522879 | Actin interacting protein 1 | |
| 2 | View Details | [733..951] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [952..1223] | 1.048941 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1224..1297] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)