













| General Information: |
|
| Name(s) found: |
hri1 /
SPAC20G4.03c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
negative regulation of translational initiation in response to osmotic stress
[IGI cellular response to stress [IMP protein amino acid phosphorylation [IDA |
| Molecular Function: |
ATP binding
[ISS protein binding [IGI eukaryotic translation initiation factor 2alpha kinase activity [NAS protein serine/threonine kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLGTSTKCSK RDCNYAKENI SRIMPTIGLG YKQNFEKKTA DTQSSCKLLL VALLESFCKH 60
61 SDQTPEQSKQ MFLYVAHSLQ NSGIIDFEFS EELEPIRNAY ADSLHNLLSK AFRSTLPMSS 120
121 TKDSKKSRYS SPDGVLAKTA SFLSVLSDGG YEDDVMNVKP SVNVLSNRLN HLPVEALESC 180
181 FPQTTLESST FADFCEHKDG TGNLSFSNFD SFPTVQRSRY ASDFEELELL GKGGYGSVYK 240
241 ARNKFDGVEY ALKKIPLRLR SFSTSSNIFR ESRTLARLNH PNVIRFFSSW VELLPSSEKQ 300
301 IEEEPLASAD ETLSQSADID NFMFDMDTGL LQHTYPSSVQ ILFQEDSVAD DLTPCYSTKN 360
361 STCNLTDLFK KEADQDYAES HDCSSTTSQV DTLGKLAPTK SASEMLLMDS FLSEREEDEC 420
421 SNIPSFDQQP LCLYIQMALC EETLEKHINR RNKHIHGVMS KGLRNCYILL FARILEGVLY 480
481 LHDAMHLVHR DLKPRNIFLS SGVHSEPCSV CLPNFSDEDN VEVSNAYEPV NQRTLCVVPK 540
541 IGDFGLVLSQ SDNLEEGTNS SAESSFVGTS TYAAPELFSK HMRSVMNNNS STDIYALGIL 600
601 FFELLYPFNT RMERASAIAN LKKGIFPHDF LDSMPEEASL IRSMLSSSNK RPTAAQLLTS 660
661 NLFHDLVVNE LHVYQALLED AEKRNNNLKA ELNILRVLNP NYDC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
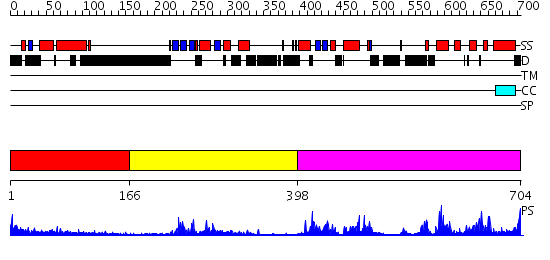
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [166..397] | 4.74 | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | |
| 3 | View Details | [398..704] | 39.221849 | Crystal Structure of eIF2alpha Protein Kinase GCN2: R794G Hyperactivating Mutant in Apo Form. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)