













| General Information: |
|
| Name(s) found: |
SPAC9G1.10c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1191 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytosol [IDA actin cortical patch [ISS |
| Biological Process: |
cellular cell wall organization
[ISS phosphatidylinositol metabolic process [IC inositol lipid-mediated signaling [IC endocytosis [ISS phosphoinositide dephosphorylation [ISS |
| Molecular Function: |
phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASRQGFSNV NEDEPELPPS VLSLKSKFES LSTGDLTNLD EKTAKRRTVK GCKNGTSEPN 60
61 VFKARPIPPP RQVSSTIGSS TGRKVSGSIQ RLASNFKNPS NPHADVSKID RLPSDSSESH 120
121 VATPSSPTIS NSFVSVSPLL KRPQQKGPEI SFQSSVQSTK GNDLMKHDDT NNHQIPPPKP 180
181 NFSSKAGSSS PISVSPLKNV KAYISQSPTH SEASSVLSSE EEEENVINSS KSVPSFDLHD 240
241 PFSQTFGKEC PISTAPPVLN IGDRSLETPP PIPSPRPPQP VAVEAIQQSR AVISQQLPLH 300
301 VSPRKPPKPP LRKVSTQRSS SPIENLATKS DASLVTGLQS SPYTHIAPAS EMSLIPEKPR 360
361 LPPRPSHTLS ELSSPALTSE NLSSKPSPLF PPPPPRVKSL ATNKPVSMPV STEQSDPSVA 420
421 ASSSSSSQLD VVLKGSIPDT SSVRRNPPCF VNGVESINVD FEARIFDVSG DRLVLAGNGG 480
481 LRVYDTVTGL CHWHMPLGDT KVTSLSFKSS PENYSDDGRF VWFGTRDGML WEVDVQNHHI 540
541 VTKKSVSNCP ITYVMVYKNE MWTLDDMGKL YVWQEDEIMG LSIQSTPHSI RTIPHATHAM 600
601 VLDNRLLWVV VGKSIYVYDP STSENESASV LAKPMTPPGL IGDISCGTTI SNFTDLVFYG 660
661 HVDGKISIFS KTQYRFLELI TSSSFYRICS LVGVGNTLWA AYTTGMIYVF DVSESPWRLL 720
721 KSWHGHKASH NGATTILGID VNSVWKAKRL QVVSMASSVV KFWDGLMMGD WLATEMRSRF 780
781 PEYSNFTDVS ILICSWNAGA SKPSDLDSDT IGASMIPMMI RDNGYPDIVV FGFQELVDLE 840
841 NKRLTARSIL SKSSSKGGSS NSANISSQYR LWREKLESEM MRVSSNDDYQ VLVCENLVGL 900
901 FSCVFVKNKL QSKIRMLQST TVKTGLGGLH GNKGAIVVRF LVDDTSYCIV NCHLAAGQSN 960
961 KAARNNDLAT ILDNASLFPE NDETDQLNTF VGGGDGSLIM DHEVCVLHGD LNYRINTLRP1020
1021 KALDLIKKND IKTLLQSDQL LVERKRNAGF RLRTFTEPEI TFAPTYKYDV HSEQYDSSEK1080
1081 KRVPAWCDRI CYRGSPDYIS AENYTRYELK ASDHRPVSAL IHSKAKMVNA QSQGSTWDVV1140
1141 KRKWIEYADE FKRKAKITYV MNYTSVSYQT AEQYLSGNNW NVQNALKQVS S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
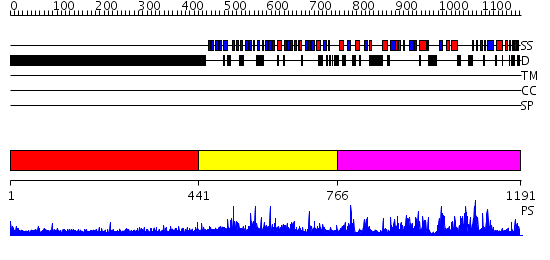
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..440] | 3.522879 | No description for 1y0fB was found. | |
| 2 | View Details | [441..765] | 6.36 | Tup1, C-terminal domain | |
| 3 | View Details | [766..1191] | 122.0 | Synaptojanin, IPP5C domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)