













| General Information: |
|
| Name(s) found: |
moc3 /
SPAC821.07c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
ascospore formation
[IMP response to calcium ion [IMP regulation of transcription, DNA-dependent [ISS cellular response to stress [IMP response to DNA damage stimulus [IMP gene-specific transcription from RNA polymerase II promoter [RCA regulation of conjugation with cellular fusion [IGI |
| Molecular Function: |
DNA binding
[ISS transcription factor activity [ISS zinc ion binding [ISS protein binding [IPI specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRNRTGSIN SNPLYIPNPN VEPTPKPTKR RTKTGCLTCR RRRIKCDETK PFCLNCTKTN 60
61 RECEGYPNSA AQMQAMGSVS PPELSVHSAQ QPLIPTSIAS SSAQTGDTFS GSSQSNFNTN 120
121 DLNQMTSSSN LSTVTPIKQD HQKPMNLQGF PSAYQQHQYL QSNHNVPTNN SSSATSSTKP 180
181 SVQSVGQASY PFLSSVSNFP SNFNSELFPF YFHDVVPSIC AFEFDNNIAL HFWSVTVPQF 240
241 AQSMPCIANS LMAFASIKKL DVFGAYSHLT RALRCPMPGP NSFEYLLVSA FLTLTQLNLP 300
301 AYDLNFCNNF IRKLSWSSST KSNYVSLLIA MVVRELVFAI LPRGCIWGFN GKPLSEVSVS 360
361 RSHAPVSDSL FTIGLDILSQ PTLSDDLHRE RMVAWRDEYH VHLYSASRSP LTKVIDCVGH 420
421 AVTKNNNDVL SGLQQLMQEE CTDIAVLRTS YLCVLSLQNV FASNSKEFKL RAQIESHFGR 480
481 LMLEHFMDCN VLNRPVL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
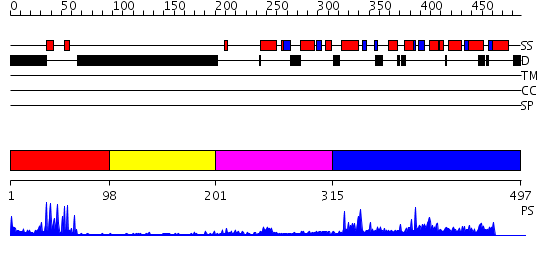
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 10.09691 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [98..200] | 2.36 | PPR1 | |
| 3 | View Details | [201..314] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [315..497] | 3.148993 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)