













| General Information: |
|
| Name(s) found: |
pli1 /
SPAC1687.05
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of chromosome segregation
[IMP protein sumoylation [IMP regulation of mitotic recombination [IMP regulation of telomere maintenance [IMP |
| Molecular Function: |
zinc ion binding
[IEA]
SUMO ligase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQANFLQEL PNVLKRLETG LIIPQLKDIL RVFGLRLSGT KAELITRIKQ LIERIAIENN 60
61 TTSWEALKKA IDGDVTSAVC ILKYNTYQIY SAAAPIAPPS SASGNRSYSR PFAPVVHSRI 120
121 RFRKSPFYDI LEQFNAPFVV PACVGTRNTI SFSFHVTPPA LSKLLNDPKQ YRVYLFSTPS 180
181 ETIGFGNCLM EFPTPQMELR INNQVAHANY RRLKGKPGTT NPADITDLVS KYAGPPGNNV 240
241 VIYYMNSTKS YSVVVCFVKV YTIENLVDQI KSRKAESKEK IIERIKNDNQ DADIIATSTD 300
301 ISLKCPLSFS RISLPVRSVF CKHIQCFDAS AFLEMNKQTP SWMCPVCASH IQFSDLIIDG 360
361 FMQHILESTP SNSETITVDP EGNWKLNTFD EPVESSEDEF VPKEKVIELS DGEGISTMAN 420
421 KSNDQPTRRA STHNSGPPAK RKRESLVIDL TISDDDENVA TSTTESPSNA TKENSLSRNV 480
481 QSPNIDTAIS NRSTNVRHGH PGFKDYTVEN SPASRERSTS ESAQSSVHMG YAGEGGLLSG 540
541 ALRAPSQQNN NNSNTQHSIN LHTIVPSPYE PPLSVTPSTA ITNLSIPESN RTNSSASSKS 600
601 FTMNDLILPP LHLKNTTQTN NAHEDAQSSN LSQNHSLFYE RIPQRPSYRI EKQNKGIYED 660
661 ENEQSISAMP IPRAHPQLPK NLLSQTAGPL WDEQQDAQVD WNSELQSNNS YHNSGFEGTG 720
721 NTFQSID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
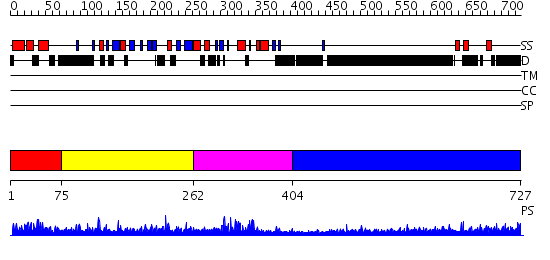
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | 16.39794 | Solution structure of human p53 binding domain of PIAS-1 | |
| 2 | View Details | [75..261] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [262..403] | 1.86 | No description for 2cklB was found. | |
| 4 | View Details | [404..727] | 1.31 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)