













| General Information: |
|
| Name(s) found: |
arg4 /
SPBC215.08c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1160 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA carbamoyl-phosphate synthase complex [ISS] |
| Biological Process: |
arginine biosynthetic process
[ISS |
| Molecular Function: |
ATP binding
[ISS manganese ion binding [IEA] carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALWATSMRR AIPGISKAFL GSNRVLETAG VSQLSKHLLP QWSGVPHRKI SASATNFNDK 60
61 VKESNTPDAN AYIRSGHIKA AEHEKVKKVV VVGSGGLSIG QAGEFDYSGA QAIKALRESS 120
121 VHTILINPNI ATIQSSHSLA DEIYFLPVTA EYLTHLIERE NPDGILLTFG GQTALNVGIQ 180
181 LDDMGVFARN HVKVLGTPIS TLKTSEDRDL FAKALNEINI PIAESVAVST VDEALQAAEK 240
241 VSYPVIIRSA YSLGGLGSGF ANNKEELQSM AAKSLSLTPQ ILVEKSLKGW KEVEYEVVRD 300
301 AANNCITVCN MENFDPLGIH TGDSIVVAPS QTLSDEEYHM LRTAAIKIIR HLGVVGECNV 360
361 QYALSPNSLE YRVIEVNARL SRSSALASKA TGYPLAYTAA KIALGHTLPE LPNAVTKTTT 420
421 ANFEPSLDYV VTKIPRWDLS KFQYVNREIG SSMKSVGEVM AVGRTFEESL QKALRQVDPS 480
481 FLGFMAMPFK DLDNALSVPT DRRFFAVVEA MLNQGYSIDK IHDLTKIDKW FLSKLANMAK 540
541 VYKELEEIGS LYGLNKEIML RAKKTGFSDL QISKLVGASE LDVRARRKRL DVHPWVKKID 600
601 TLAAEFPAHT NYLYTSYNAS SHDIDFNEHG TMVLGSGVYR IGSSVEFDWC GVSCARTLRK 660
661 LGHSTIMVNY NPETVSTDFD ECERLYFEEL SYERVMDIYE METASGIVVS VGGQLPQNIA 720
721 LKLQETGAKV LGTDPLMIDS AEDRHKFSQI LDKIGVDQPA WKELTSVAEA SKFANAVGYP 780
781 VLVRPSYVLS GAAMSVIRDE SSLKDKLENA SAVSPDHPVV ITKFIEGARE LDVDAVASKG 840
841 KLLVHAVSEH VENAGVHSGD ATIALPPYSL SEDILSRCKE IAEKVCKAFQ ITGPYNMQII 900
901 LAQNPDKPDT PDLKVIECNL RASRSFPFVS KTLGVNFIDV ATRSIIDQEV PAARDLMAVH 960
961 RDYVCVKVPQ FSWTRLAGAD PYLGVEMSST GEVACFGKDV KEAYWAALQS TQNFKIPLPG1020
1021 QGILLGGDRP ELAGIAADLS KLGYKLYVAN KDAAKLLQPT SAEVVEFPVK DKRALRAIFE1080
1081 KYNIRSVFNL ASARGKNVLD QDYVMRRNAV DFNVTLINDV NCAKLFVESL KEKLPSVLSE1140
1141 KKEMPSEVKR WSEWIGSHDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
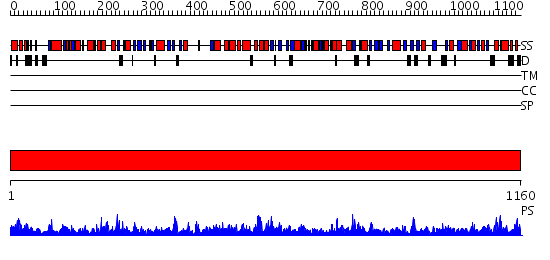
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1160] | 1000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)