













| General Information: |
|
| Name(s) found: |
SPAC105.03c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS fungal-type vacuole [IDA |
| Biological Process: |
gene-specific transcription from RNA polymerase II promoter
[RCA regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
DNA binding
[ISS transcription factor activity [ISS zinc ion binding [ISS specific RNA polymerase II transcription factor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQSSSPSQP ESQAQPNLCI PCSLGTCLIH FHEFSSSSFM DPVSLFCSSP YPNLPPHSRS 60
61 SSLESKKPSV ASQDVKSDGT LPIGTNNNPL IPSHSQESSH WTIRHESMPS ALAGSSAQSM 120
121 QQFPSITQNE ENFRFKKSFT QPQPIVKETT FPKSEPGQEH AKLSDLSYEE FLKKYSSTKV 180
181 ERVSEAAPPP SSLNSSTVLD ENDSLISQGS SVDDQTDFLG FDDSLSYAYI LNPTSDSDVD 240
241 LIRQYFIPKE GTYTFSNMHI RYVSANPKTP VMYMVDPAYE PHKQIARYEK LQQIFQVVEA 300
301 TVSPEVGERL IGLYFRYIHR VYPVIHGERF LLAFKKAKHK LSPILLTAMY ASSIIYWDAD 360
361 KELRNYPRLD AKKLWDLTED FLDLSFSLSR LSTLQAAIIF LTGRPWINVA GNWSILTRAI 420
421 ALAQILGFHL DCSEWQIPKE EKILRNRVWW ALFVNEKWLS MYIGINASIR QDDYLVPPLT 480
481 DNQLLAGEQR IADSFNVFVK MAELSILLQN VLQHLFNARA VMTMSKNRRT VAQKISKYLI 540
541 SLEEFSCSCN LRENQPGSLS LFLQIDALEL LLRKTALKLK LTDAVYQQEM LNVVQRCVSH 600
601 FLAINESHFT DFFWPYSQFY FCILSSAIIK MYLDFEFNEN FNATTFGLLS SFTKHCVWLK 660
661 FRNFDLVFMA YKRLNALLIE LSSGRPLIGK LLQETFEHRK RNLDETLT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
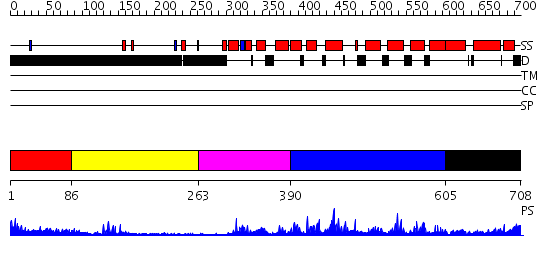
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 1.252997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [86..262] | 2.13 | PPR1 | |
| 3 | View Details | [263..389] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [390..604] | 5.105996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [605..708] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)