













| General Information: |
|
| Name(s) found: |
mst1 /
SPAC637.12c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 463 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IDA NuA4 histone acetyltransferase complex [IDA |
| Biological Process: |
histone H4 acetylation
[ISS centromere complex assembly [IGI chromatin remodeling [RCA replication fork protection [IGI chromosome segregation [IMP chromatin assembly or disassembly [IEA] regulation of transcription [IEA] double-strand break repair [IGI |
| Molecular Function: |
protein binding
[IPI chromatin binding [IEA] H4 histone acetyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNDVDDESK IETKSYEAKD IVYKSKVFAF KDGEYRKAEI LMIQKRTRGV VYYVHYNDYN 60
61 KRLDEWITID NIDLSKGIEY PPPEKPKKAH GKGKSSKRPK AVDRRRSITA PSKTEPSTPS 120
121 TEKPEPSTPS GESDHGSNAG NESLPLLEED HKPESLSKEQ EVERLRFSGS MVQNPHEIAR 180
181 IRNINKICIG DHEIEPWYFS PYPKEFSEVD IVYICSFCFC YYGSERQFQR HREKCTLQHP 240
241 PGNEIYRDDY ISFFEIDGRK QRTWCRNICL LSKLFLDHKM LYYDVDPFLF YCMCRRDEYG 300
301 CHLVGYFSKE KESSENYNLA CILTLPQYQR HGYGKLLIQF SYELTKREHK HGSPEKPLSD 360
361 LGLISYRAYW AEQIINLVLG MRTETTIDEL ANKTSMTTND VLHTLQALNM LKYYKGQFII 420
421 CISDGIEQQY ERLKNKKRRR INGDLLADWQ PPVFHPSQLR FGW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
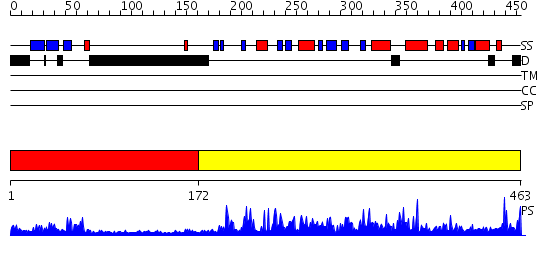
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 16.69897 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [172..463] | 126.0 | Histone acetyltransferase ESA1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)