













| General Information: |
|
| Name(s) found: |
lys2 /
SPAC343.16
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 721 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
generation of precursor metabolites and energy
[RCA glutamate metabolic process [RCA lysine biosynthetic process via aminoadipic acid [IMP |
| Molecular Function: |
iron ion binding
[IEA]
homoaconitate hydratase activity [IMP 4 iron, 4 sulfur cluster binding [IEA] iron-sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSGEMHHPY QAFSKVGKCE ISQTNPSFSS GMRCLVRSAD IQFKGICGLT RGFASFNKPP 60
61 QTITEKIVQK FAQNIPENKY VRSGDYVTIK PKHCMSHDNS WPVALKFMGI GAKKVFDNRQ 120
121 IVCTLDHDVQ NKSEANLRKY KNIESFAKGQ GIDFYPAGRG IGHQIMVEQG YAMPGSMAVA 180
181 SDSHSNTYGG VGCLGTPIVR TDAAAIWATG QTWWQIPPIA RVNLVGQLPK GLSGKDIIVS 240
241 LCGAFNHDEV LNHAIEFYGE GLNSLSIESR LTIANMTTEW GALSGLFPTD EKLLAWYEDR 300
301 LKFLGPNHPR VNRETLDAIK ASPILADEGA FYAKHLILDL STLSPAVSGP NSVKVYNSAA 360
361 TLEKKDILIK KAYLVSCTNG RLSDIHDAAE TVKGKKVADG VEFYVGAASS EVEAAAQKNG 420
421 DWQTLIDSGA RTLPAGCGPC IGLGTGLLKD GEVGISATNR NFKGRMGSRE ALAYLASPAV 480
481 VAASAIAGKI VAPEGFKNAV SLVSAVDITD KVNKQTASKS STEAVDSETA IIDGFPSIVA 540
541 GEIVFCDADN LNTDGIYPGR YTYRDDITKE EMAKVCMENY DSEFGKKTKK DDILVSGFNF 600
601 GTGSSREQAA TAILSRGIPL VVGGSFSDIF KRNSINNALL AIQLPDLVQK LRTAFANESK 660
661 ELTRRTGWHL KWDVRKSTVT VTTSDNKEMS WKIGELGNSV QSLFVRGGLE GWVKHEISKS 720
721 N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
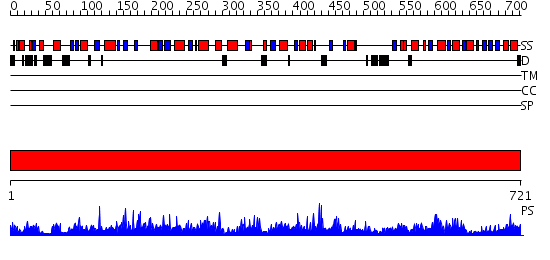
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..721] | 1000.0 | STRUCTURE OF ACTIVATED ACONITASE. FORMATION OF THE (4FE-4S) CLUSTER IN THE CRYSTAL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)