













| General Information: |
|
| Name(s) found: |
SPAC19G12.08
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 347 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IC membrane [ISS integral to membrane [IEA] endoplasmic reticulum [IDA |
| Biological Process: |
fatty acid oxidation
[IC fatty acid metabolic process [ISS sphingolipid metabolic process [ISS |
| Molecular Function: |
iron ion binding
[IEA]
sphingosine hydroxylase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVTSEKCV ILSDGTEYDV TNYLVANKDA ADLLRRYHRQ EVADILNATS KSKHSEAVVE 60
61 ILKSAKVPLK NKEFSDLVDQ NIGVGYGNEF IVKPTDLDKD FEKNHFLDLK KPLLPQILFG 120
121 NIKKDVYLDQ VHRPRHYRGS GSAPLFGNFL EPLTKTPWYM IPLIWVPCVT YGFLYACTGI 180
181 PFSVAITFFI IGLFTWTLVE YTMHRFLFHL DEYTPDHPIF LTMHFAFHGC HHFLPADKYR 240
241 LVMPPALFLI FATPWYHFIQ LVLPHYIGVA GFSGAILGYV FYDLTHYFLH HRRMPNAYLT 300
301 DLKTWHLDHH YKDYKSAYGI TSWFWDRVFG TEGPLFNEQG KISTKAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
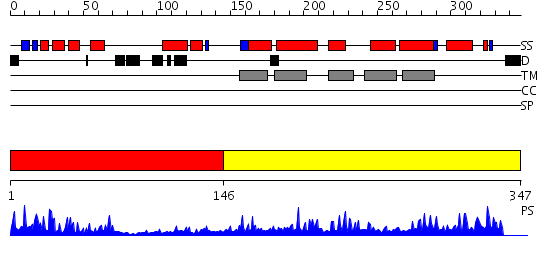
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 14.69897 | Sulfite oxidase, C-terminal domain; Sulfite oxidase, N-terminal domain; Sulfite oxidase, middle catalytic domain | |
| 2 | View Details | [146..347] | 17.136677 | No description for PF04116.4 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| Source: Reynolds et al. 2008. Manuscript submitted |

|