













| General Information: |
|
| Name(s) found: |
mug30 /
SPBP8B7.27
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA cell tip [IDA spindle pole body [IDA |
| Biological Process: |
protein ubiquitination
[RCA |
| Molecular Function: |
zinc ion binding
[IEA]
small conjugating protein ligase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRPIEIANV SSGNRRTLPA LHGSSFSSRS ELDNNTNSKI SRRLRELVCQ RMGENPLRLR 60
61 QSFHETISND SYGNSEKLVI RPCICCNSVL RYPAQAMCFR CSLCMTVNDV YFCLSGSQIK 120
121 TKASTDGSQF ISVEHFVETI HQTRSALQLA KQRTGNVQAI VAAGLPLREL ISLVFGSPPI 180
181 LNKLFSVKNG CSIQSSGLNY KLIYQLYHDI TNLDILITNE LLRAIESLLR RPMLYCHDPA 240
241 DYQYLLILLE NPLLNSKSKN IVNKSSSILK RILGVLSNLN EKTHHFFISC FKKQPYNNPK 300
301 FFRRKVDLIN KFIGQRLMET YSRNKRKHYY NNDWQIKSAA ITMALLYSAN SQMRLIDRSS 360
361 FYCIMADFIN LYHDFELWEQ KINCFCFCQY PFLLSMGAKI SILQLDARRK MEIKAREAFF 420
421 SSILSKMNVE PYLMIRVRRD RLLEDSLRQI NDRNKDFRKA LKVEFLGEEG IDAGGLKREW 480
481 LLLLTRKVFS PEFGLFVNCE ESSNYLWFNY SHRSKEIDYY HMSGILMGIA IHNSINLDVQ 540
541 MPRAFYKKLL QLPLSFNDLD DFQPSLYRGL KELLLFEGDV KNTYGLNFTI NLKAVEGFRT 600
601 VELKEGGSEL SVDNENRKEY VLRYVDYLLN TTVKKQFSAF FDGFMKVCGG NAISLFQDNE 660
661 ISKLIRGSEE VIDWELLKNV CVYDFYDQNA ISNSISESEP SMTASKYLCH SFVSKRKIIL 720
721 WFWDLISHYS LKMQKLFLIF VTGSDRIPAT GAHNFQLRIS VLGPDSDQLP ISHTCFNHLC 780
781 IWEYSSREKL KKKLDTALLE TNGFNIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
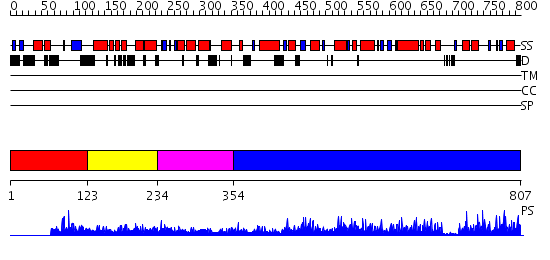
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 1.06 | CREB-binding transcriptional adaptor protein CBP (p300) | |
| 2 | View Details | [123..233] | 3.076989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [234..353] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [354..807] | 110.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)